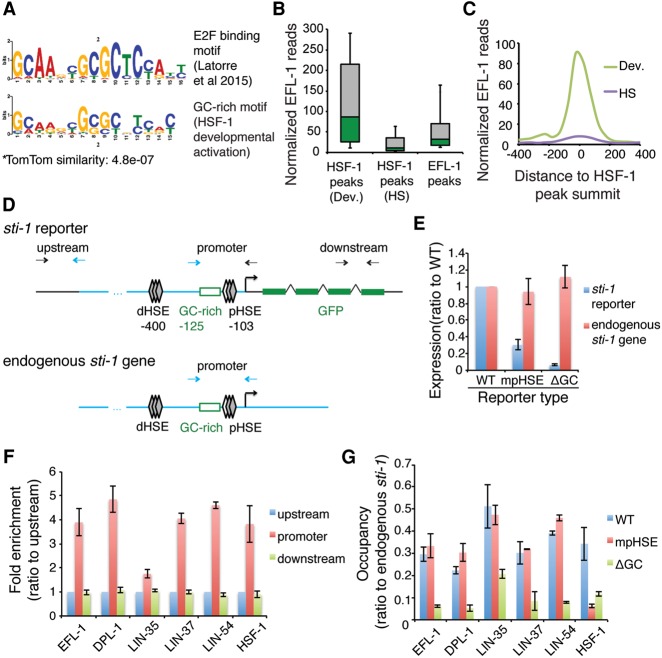
Figure 5.
An E2F complex binds to the GC-rich motif at HSF-1 developmental genes. (A) The GC-rich motif from HSF-1 directly activated promoters in development (bottom) resembles an E2F-binding motif (top) derived from ChIP-seq peaks of the E2F-associated DRM complex (Latorre et al. 2015). (B) Box plots of EFL-1 ChIP-seq reads at HSF-1 peaks that are linked to either developmental activation (Dev.), heat-shock induction (HS), or all EFL-1 peaks at promoters (±250 bp around summits) (Kudron et al. 2013). Peaks linked to HSF-1 activation in both conditions were excluded. Boxes depict the 25th through 75th percentiles, and whiskers show the 10th through 90th percentiles. (C) Composite plots of EFL-1 ChIP-seq reads (Kudron et al. 2013) within 400 bp from the summits of the HSF-1 peaks that are linked to either developmental activation (Dev.) or heat-shock induction (HS). (D) Schematic representation of ChIP-qPCR analysis at the integrated sti-1 reporters. Paired arrows represent the primers used in qPCR analysis, blue lines and arrows indicate the sequences from the endogenous sti-1 promoter, and black lines and arrows indicate the sequences specific to the reporter transgene. (E) RT-qPCR analysis of the sti-1 reporter and the endogenous sti-1 gene in transgenic animals. Normalized expression levels of the reporter (GFP/mCherry) and the endogenous gene (sti-1/housekeeping genes) were measured in transgenic animals carrying the sti-1 reporter containing the wild-type (WT) promoter or promoter variants with mutations in the proximal HSE (mpHSE) or deletion of the GC-rich motif (ΔGC). Data are represented as ratios of the expression levels in animals carrying the wild-type reporter. Error bars represent the SEM of biological triplicates. (F) ChIP-qPCR analysis of HSF-1 and subunits of the DRM complex in transgenic animals carrying the wild-type sti-1 reporter. Occupancies of the transcription factors at three regions across the reporter were measured and are shown as fold enrichment over the upstream control region. Error bars represent the SEM of biological triplicates. (G) ChIP-qPCR analysis of HSF-1 and subunits of the DRM complex in transgenic animals carrying different sti-1 reporters. Transgenic animals with an integrated sti-1 reporter carrying the wild-type (WT) promoter, mutation of the proximal HSE (mpHSE), or deletion of the GC-rich motif (ΔGC) were analyzed. Occupancies of the transcription factors were measured at the promoter regions of the reporter as well as the endogenous sti-1 gene, and the normalized occupancy (reporter/endogenous gene) is shown in the histograms. Error bars represent the SEM of biological triplicates.