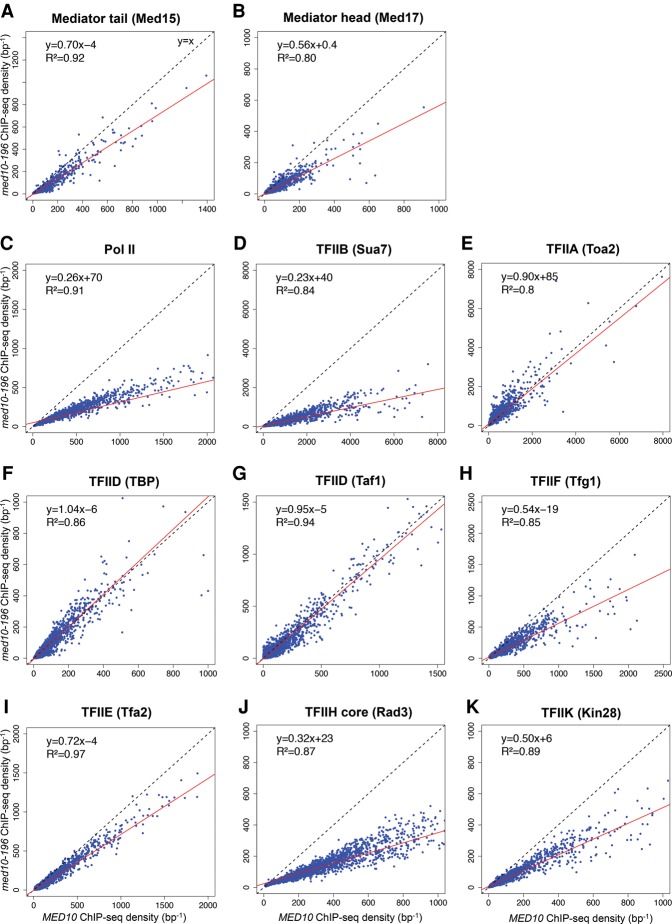
Figure 4.
Genome-wide effects of the med10-196 mutation on PIC component distribution. Cells were grown at 30°C in YPD medium and then transferred for 90 min to 37°C. The ChIP-seq densities of sequence tags in Mediator tail (Med15) (A), Mediator head (Med17) (B), TFIIB (Sua7) (D), TFIIA (Toa2) (E), TFIID (TBP) (F), TFIID (Taf1) (G), TFIIF (Tfg1) (H), TFIIE (Tfa2) (I), TFIIH core (Rad3) (J), and TFIIK (Kin28) (K) were calculated for promoter regions of Pol II transcribed genes. (C) The densities of sequence tags in the Pol II ChIP-seq experiments were calculated for the Pol II transcribed genes. Tag densities were normalized relative to qPCR data on a set of selected genes. Each point on the plot corresponds to one promoter region or one ORF. Promoter regions correspond to intergenic regions in tandem or in divergent orientation, excluding intergenic regions encompassing Pol III transcribed genes. In all, 2694 intergenic regions corresponding to 3303 Pol II-enriched genes were used for these analyses. A linear regression (red line) for ChIP-seq density in the mutant versus ChIP-seq density in wild type and an R2 correlation coefficient are indicated.