Abstract
The glycine riboswitch often occurs in a tandem architecture, with two ligand-binding domains (aptamers) followed by a single expression platform. Based on previous observations, we hypothesized that “singlet” versions of the glycine riboswitch, which contain only one aptamer domain, are able to bind glycine if appropriate structural contacts are maintained. An initial alignment of 17 putative singlet riboswitches indicated that the single consensus aptamer domain is flanked by a conserved peripheral stem–loop structure. These singlets were sorted into two subtypes based on whether the active aptamer domain precedes or follows the peripheral stem–loop, and an example of each subtype of singlet riboswitch was characterized biochemically. The singlets possess glycine-binding affinities comparable to those of previously published tandem examples, and the conserved peripheral domains form A-minor interactions with the single aptamer domain that are necessary for ligand-binding activity. Analysis of sequenced genomes identified a significant number of singlet glycine riboswitches. Based on these observations, we propose an expanded model for glycine riboswitch gene control that includes singlet and tandem architectures.
Keywords: riboswitch, glycine, aptamer, gene control, metabolite
INTRODUCTION
The glycine riboswitch plays an important role in bacterial glycine homeostasis and offers an unusual example of gene regulation due to its unique tandem architecture (Barrick and Breaker 2007; Barrick et al. 2004; Mandal et al. 2004; Kazanov et al. 2007; Lipfert et al. 2007; Kwon and Strobel 2008; Huang et al. 2010; Butler et al. 2011; Erion and Strobel 2011; Kladwang et al. 2012; Sherman et al. 2012; Baird and Ferré-D'Amaré 2013; Esquiaqui et al. 2014; Ruff and Strobel 2014; Zhang et al. 2014). Glycine riboswitches control genes related to the import, synthesis, and degradation of glycine, and these genes maintain the intracellular glycine concentration within a narrow range in bacterial cells (Barrick and Breaker 2007; Barrick et al. 2004; Kazanov et al. 2007). Disruption of the riboswitches has been shown to be toxic to S. griseus growing in media containing glycine (Tezuka and Ohnishi 2014). More than 7000 examples of the riboswitch class are found across bacterial phyla and environmental sequences (P McCown and R Breaker, unpubl.).
Previous sequence analysis of the glycine riboswitch identified a tandem structure, with two homologous ligand-binding domains (aptamers) that each binds a separate glycine molecule (Mandal et al. 2004). Unlike the few other known examples of tandem riboswitches, the two aptamer domains are followed by a single expression platform (Mandal et al. 2004; Sudarsan et al. 2006; Welz and Breaker 2007; Breaker 2011). Because the two tandem aptamers have been conserved against evolutionary drift, they are expected to provide some benefit relative to the simpler, single-aptamer riboswitch. It was originally proposed that the two aptamer domains are cooperative, providing a sharper, more digital response to changes in glycine concentration (Mandal et al. 2004; Lipfert et al. 2007; Kwon and Strobel 2008; Erion and Strobel 2011). Recent work has shown that, at least for in vitro binding assays, certain glycine riboswitch constructs examined under equilibrium conditions are not cooperative (Kladwang et al. 2012; Sherman et al. 2012; Baird and Ferré-D'Amaré 2013), and the two binding sites bind ligand independently (Ruff and Strobel 2014). Further research is necessary to determine whether tandem riboswitches act cooperatively in vivo.
The two aptamer domains of the tandem riboswitch interact to form a network of conserved structural contacts (Fig. 1A,B; Huang et al. 2010; Butler et al. 2011). Biochemical analysis has shown that these tertiary interactions between the aptamer domains are necessary for ligand-binding activity and that ligand binding and interface formation are linked equilibria (Erion and Strobel 2011; Ruff and Strobel 2014). The two aptamer domains are connected by a kink-turn and P0 helix (Kladwang et al. 2012; Sherman et al. 2012), which likely stabilize and organize the two binding domains. We have previously hypothesized that the first aptamer domain could be important for stabilizing or kinetically scaffolding the second (Ruff and Strobel 2014), whose formation is proposed to be important for gene control (Mandal et al. 2004).
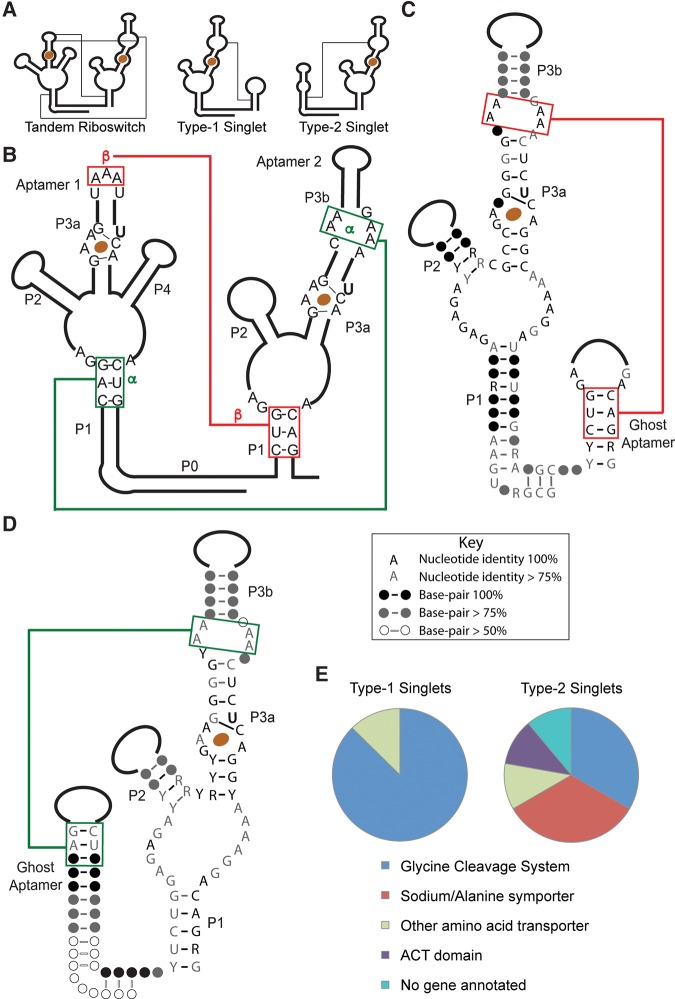
FIGURE 1.
Consensus sequences and distribution of singlet glycine riboswitches based on an initial alignment of 17 sequences. (A) Schematic diagrams showing the secondary structures and tertiary interactions of tandem, type-1 singlet, and type-2 singlet riboswitches. (B–D) Consensus sequences for tandem (B), type 1 singlet (C), and type 2 singlet riboswitches (D). The α and β A-minor motifs are conserved. (E) Distribution of genes controlled by the singlet riboswitches in the initial alignment.
The original alignment of glycine riboswitches identified a small number that contain only a single-aptamer domain (Mandal et al. 2004). Each subsequent alignment has identified a small percentage of these “singlet” riboswitches (Kazanov et al. 2007; Kladwang et al. 2012; P McCown and R Breaker, unpubl.), but these sequences have not been characterized. Given the importance of aptamer dimerization in the tandem system (Ruff and Strobel 2014), we set out to determine if singlet versions of the glycine riboswitch are able to bind glycine and are likely to serve as genetic switches. We biochemically characterized two prototypical examples, showing that the singlet aptamers do bind ligand and are dependent on a “ghost-aptamer” domain for ligand-binding activity. Based on these observations, we propose an expanded model for glycine riboswitch control of gene expression that incorporates singlet riboswitch activity.
RESULTS
Putative glycine singlet aptamers are flanked by conserved stem–loop “ghost aptamers”
Seventeen putative singlet riboswitches were initially identified by inspection in searches for tandem riboswitches (Supplemental Fig. S1; J Barrick and R Breaker, unpubl.). Because this initial set of singlet riboswitches appeared in searches for tandem switches even while missing one aptamer, their sequences are closely related to that of the consensus glycine aptamer. The single aptamer domains are flanked by adjacent conserved helical elements with the sequence and length of the P1 stem of a glycine aptamer domain (consensus sequences shown in Fig. 1C,D). In addition, the P0 and kink-turn motifs are conserved, indicating that the helical domain is positioned in the same orientation as a P1 stem with respect to the extant aptamer domain. Although the helical sequence is conserved, the loop length and sequence vary widely among the singlets. There is no evidence of a ligand-binding site and none of the conserved elements of a glycine aptamer are retained, except the P1 stem. We therefore dubbed these helical regions “ghost aptamers.” The singlets can be classified into two subtypes based on whether the aptamer domain precedes the ghost aptamer (type-1) or follows it (type-2). These subtypes are relatively evenly distributed among the small number of riboswitches in the initial alignment. Interestingly, all of the type-1 singlet riboswitches are predicted to be “ON” switches, while the type-2 examples have less obvious expression platforms but are likely a mixture of “ON” and “OFF” switches.
The top three base pairs (bp) of the P1 stem of a glycine aptamer in a tandem riboswitch have been shown to participate in extensive A-minor tertiary interactions with A-rich bulges in the P3 stem of the partner aptamer (Fig. 1A). Because the stem length and the sequence of these three base pairs are conserved in the ghost aptamers of singlet riboswitches, we hypothesized that they might fulfill a similar structural role in the singlet architecture. Sequence analysis of the relevant bulge of the singlet aptamer domain indicates that the adenines are conserved, and the formation of A-minor interactions is possible (Fig. 1A–D).
The P1 stem of the first aptamer in a tandem riboswitch is considerably more stable than that of the second aptamer (8–9 bp vs. 5–6 bp) (Ruff and Strobel 2014). This stability difference is also observed in the singlet riboswitches, with the stem of the ghost aptamer more stable in type-1 singlets, and less stable in type-2 singlets. This difference has implications for the gene-control model, as discussed below.
Type-1 and type-2 singlet riboswitches bind glycine as tightly as tandem riboswitches and are active as monomers
Prototypical examples of each type of singlet were chosen for biochemical analysis in vitro. A type-1 singlet from Listeria monocytogenes (Lmo) and a type-2 singlet from Desulfitobacterium hafniense (Dha) were synthesized by in vitro transcription, and equilibrium dialysis was used to determine the glycine-binding affinity. Both bind glycine with low micromolar affinities (Table 1; Fig. 2), comparable to that of the tandem riboswitch from Vibrio cholerae (VC) (Kladwang et al. 2012; Sherman et al. 2012; Baird and Ferré-D'Amaré 2013; Ruff and Strobel 2014).
TABLE 1.
Ligand-binding affinities of singlet glycine riboswitches
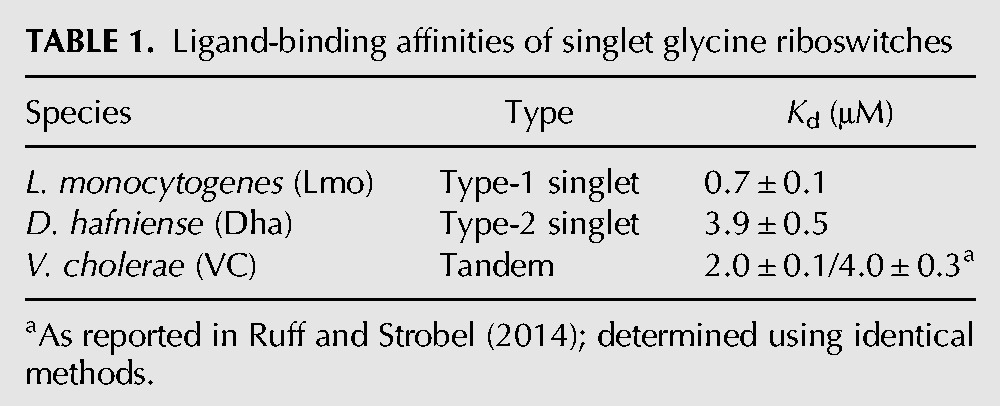
FIGURE 2.
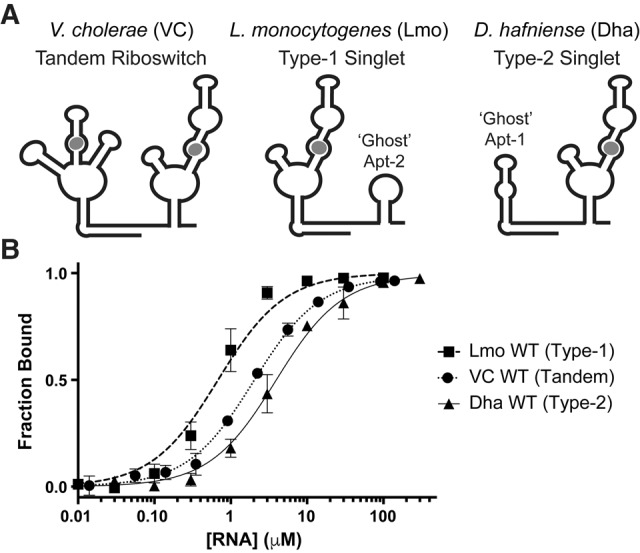
Singlet glycine riboswitches bind glycine as strongly as tandem riboswitches. (A) Diagrams of prototypical tandem (VC), type-1 singlet (Lmo), and type-2 singlet (Dha) riboswitches. (B) Glycine-binding curves for prototypical singlet and tandem riboswitches. VC WT is as reported in Ruff and Strobel (2014).
Isolated glycine aptamer domains can homodimerize in vitro (Huang et al. 2010; Sherman et al. 2012; Ruff and Strobel 2014), but only monomeric binding activity is expected to be relevant to the in vivo function of the singlet riboswitch. The singlet riboswitches were analyzed by size-exclusion fast performance liquid chromatography (FPLC) and multi-angle light scattering (MALS) to determine their multimeric state. The Lmo type-1 singlet is >95% monomer when refolded in the presence of saturating ligand (Supplemental Fig. S3). The Dha type-2 singlet is 70%–80% monomer when refolded in the presence of saturating ligand. We isolated >90% monomeric wild-type Dha (WT) using a native preparative-FPLC method and determined that it bound to glycine with similar affinity to that of the monomer/dimer mixture. The monomer was stable at room temperature overnight (16 h), or for several days at 4°C. Based on these analyses, we conclude that the Lmo type-1 and Dha type-2 singlets are able to bind glycine as monomers.
Singlet riboswitches have a single ligand-binding site that is analogous to those found in tandem riboswitches
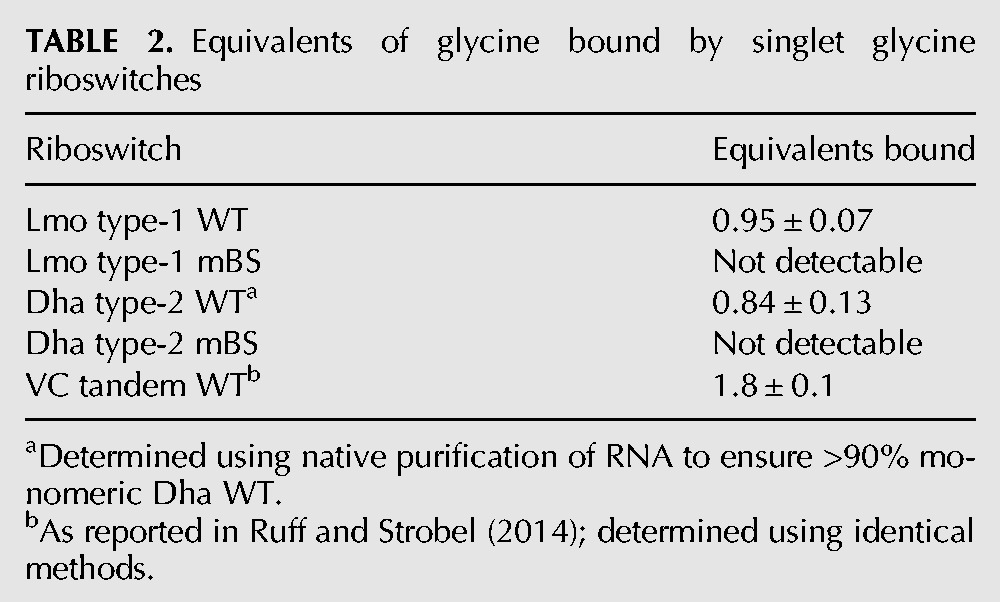
The extant aptamer domains of singlet glycine riboswitches contain the conserved ligand-binding-site sequence found in tandem riboswitches, but it is missing in the peripheral “ghost-aptamer” sequences (Figs. 1, 3). Therefore, singlet riboswitches are expected to bind only a single molecule of glycine. We determined the equivalents of glycine bound per singlet RNA molecule using equilibrium dialysis with a known excess of glycine and found that both Lmo type-1 and Dha type-2 singlets bind one equivalent of glycine (Table 2).
FIGURE 3.
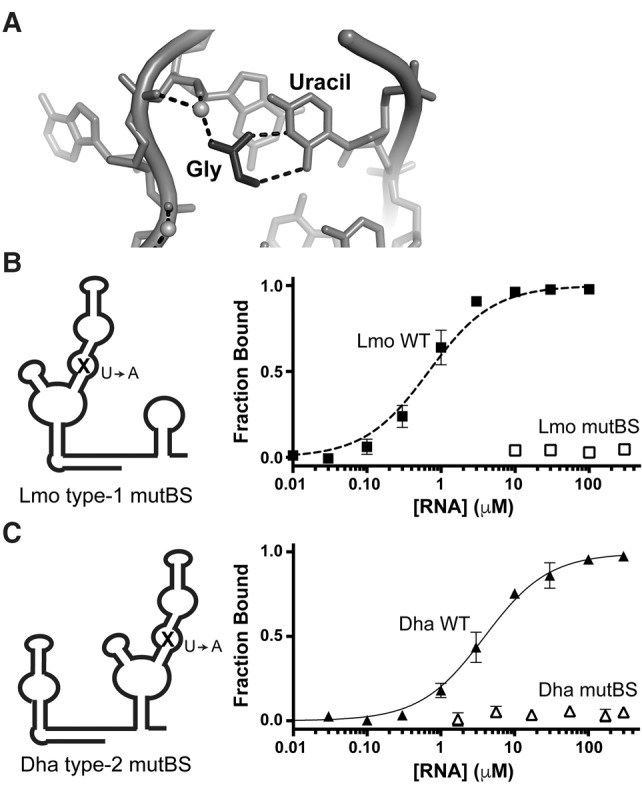
Singlet riboswitches bind glycine with a canonical binding-site motif. (A) Structural model of one glycine-binding site from the F. nucleatum tandem riboswitch (Butler et al. 2011), showing the conserved uracil that contacts glycine (black). (B,C) Diagrams depicting binding-site mutants and glycine-binding curves for wild-type (closed symbols) and mutated binding sites (open symbols), demonstrating that the singlet binding sites can be disrupted with canonical U-to-A mutations.
TABLE 2.
Equivalents of glycine bound by singlet glycine riboswitches
Mutating the uracil (U) that contacts the glycine (Fig. 3) to an adenine (A) disrupts ligand binding in the VC tandem riboswitch (Ruff and Strobel 2014). We incorporated this mutation into the binding site of each singlet (see Supplemental Material for full sequences) and determined the ligand-binding affinity of the binding-site mutants (mutBS). Both Lmo type-1 and Dha type-2 binding-site mutants show no detectable glycine binding at the highest RNA concentrations tested (Tables 2, 3; Fig. 3). These data demonstrate that singlet riboswitches contain a single canonical ligand-binding site within their single aptamer domain.
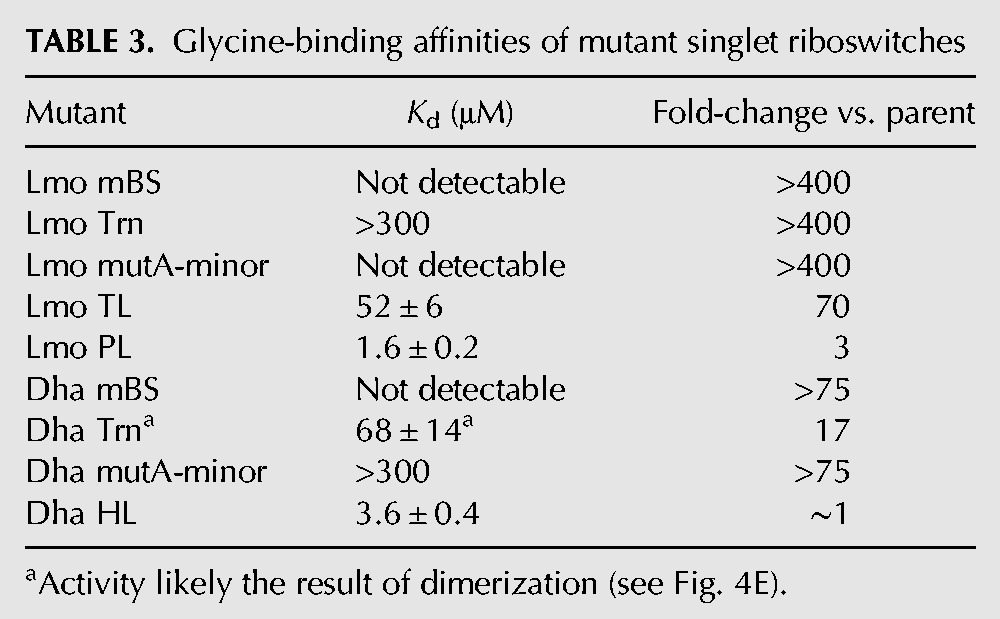
TABLE 3.
Glycine-binding affinities of mutant singlet riboswitches
Ghost aptamers mediate proper folding and ligand-binding activity of singlet riboswitches via formation of A-minor interactions with the aptamer domains
Based on the conservation of the sequence at the top of the helices of the ghost aptamers (Figs. 1C,D, 4A) and the similarity of this sequence to that found in the tandem riboswitch, we tested if these regions form similar tertiary contacts with the aptamer domains. We deleted the entire ghost aptamer (Lmo type-1 singlet) or the conserved base pairs at the top of the stem (Dha type-2 singlet) and determined the ligand-binding affinity of the resulting truncation mutants using equilibrium dialysis. Deletions were chosen to preserve the kink-turn and P0 helix in an attempt to minimize dimerization (see Supplemental Material for full sequences).
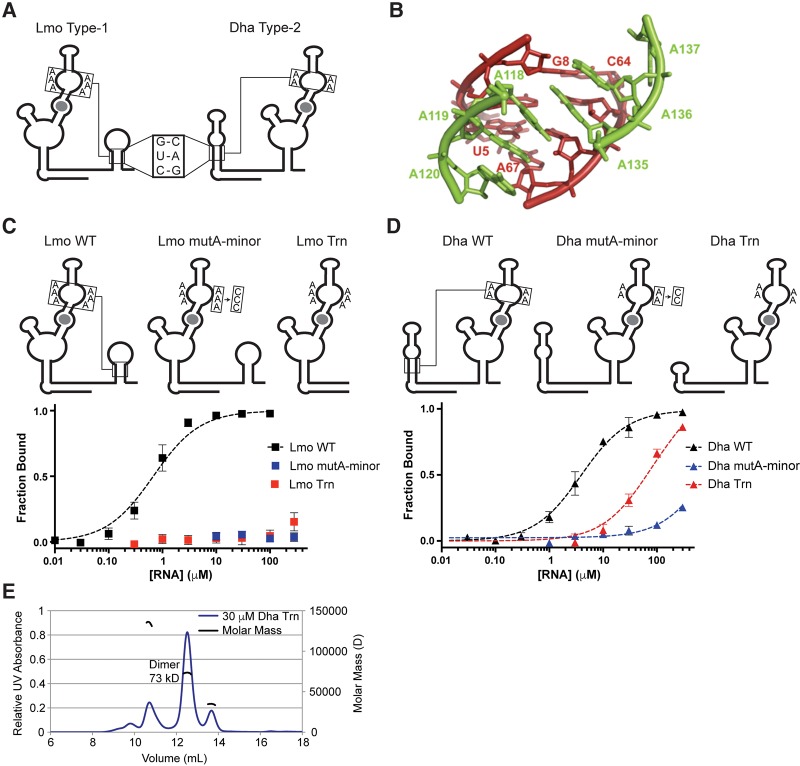
FIGURE 4.
Singlet riboswitches require conserved A-minor tertiary interactions with an adjacent “ghost-aptamer” domain. (A) Diagrams highlighting the conserved helical regions and A-rich bulges in the Lmo and Dha riboswitches. (B) Structural model of the conserved A-minor interaction from the F. nucleatum tandem riboswitch (Butler et al. 2011). (C,D) Diagrams and glycine-binding curves for singlet mutants with disrupted A-minor interactions. (E) MALS analysis of Dha Trn refolded in the presence of saturating glycine, showing that the major species is an alternative dimer conformation.
The Lmo type-1 truncation mutant (Trn) shows barely detectable binding at the highest RNA concentration tested (Table 3; Fig. 4C), indicating a greater than 400-fold disruption of ligand-binding affinity. The Dha type-2 truncation mutant binds glycine with modest affinity (KD = 68 µM), which is 17-fold weaker than Dha WT (Table 3; Fig. 4D). However, this ligand-binding activity likely depends on spurious dimerization of the Dha Trn construct (see Results). Therefore, in both type-1 and type-2 singlets, the conserved ghost aptamers are required for high-affinity monomeric binding.
We next sought to determine if the flanking helical regions form A-minor interactions with the aptamer domains. Structural (Butler et al. 2011) and biochemical (Kwon and Strobel 2008; Erion and Strobel 2011; Ruff and Strobel 2014) characterizations of tandem riboswitches have shown that A-minor interactions form between A-rich bulges in one aptamer and the P1 stem of the other aptamer (Fig. 4B) and that these interactions are important for high-affinity binding. Singlet riboswitches contain conserved A-rich bulges in their aptamer domains (Figs. 1, 4A), and these may form similar A-minor interactions with the ghost-aptamer helices. We mutated the adenines to cytosines (Doherty et al. 2001; Kwon and Strobel 2008; Erion and Strobel 2011; Ruff and Strobel 2014) in one-half of the A-rich bulge for each singlet and determined the effect on ligand-binding affinity using equilibrium dialysis.
The Lmo type-1 A-minor mutant construct shows no detectable binding at the highest RNA concentration tested, again indicating at least a 400-fold disruption of ligand-binding affinity (Table 3; Fig. 4C). The Dha type-2 A-minor mutant construct shows barely detectable binding at the highest concentration, a greater than 75-fold disruption of ligand-binding affinity (Table 3; Fig. 4D). The magnitudes of these effects are greater than the 10- to 30-fold effects seen for A to C mutations in the VC system (Ruff and Strobel 2014). Therefore, the A-minor tertiary interactions formed with the peripheral stem–loop domains are necessary for proper folding and high-affinity binding by singlet riboswitches.
Unexpectedly strong ligand binding by the Dha truncation mutant occurs in an alternative dimeric conformation
The Dha type-2 truncation mutant is the only one of the four singlet constructs containing mutated tertiary interactions that binds glycine with even moderate affinity (Table 3). Given the known tendency of isolated aptamer-2 domains to homodimerize (Huang et al. 2010; Sherman et al. 2012; Ruff and Strobel 2014), we questioned if the observed binding affinity resulted from spurious dimerization of the Dha truncation mutant construct in vitro. The Dha type-2 wild-type and mutant constructs were analyzed by size-exclusion FPLC to determine their multimeric state in the presence and absence of ligand (Supplemental Fig. S4). The Dha truncation mutant was further characterized by MALS to confirm its dimerization in the presence of glycine (Fig. 4E).
Because a significant portion of the Dha Trn pool exists as dimer across the RNA concentrations where ligand binding occurs (Fig. 4E; Supplemental Fig. S4), and because the dimer binds ligand more tightly than monomer (see Supplemental Material), we expect that the ligand-binding activity of Dha Trn is due to a spurious dimer state. The multimeric states of the other Dha mutants were also analyzed by FPLC (Supplemental Fig. S4). All form some dimers, particularly at the highest RNA concentrations tested, but in no other case did ligand binding promote formation of the dimer. Therefore, the observed ligand-binding affinities of the other Dha mutants are attributed to monomeric states.
The nonconserved loops and helical extensions of the ghost aptamers are not necessary for ligand-binding activity
Although the helical regions of the ghost aptamers are conserved in length and sequence, the loops vary widely. We tested if these variable regions could be deleted without affecting binding affinity. Lmo type-1 WT contains a large 20-base loop, with some possible base-pairing within the loop. We replaced this large loop with a pentaloop (PL) containing the sequence of the S. mutans ghost aptamer, the smallest loop sequence observed in our alignment. This mutation largely retains the wild-type ligand-binding affinity, indicating that the Lmo loop sequence is unimportant for singlet folding or binding activity. Further truncation to a GNRA tetraloop (TL) causes a 70-fold loss of ligand-binding affinity (Table 3; Fig. 5A). The tetraloop may constrain the top of the helix in such a way that it disrupts the A-minor interactions.
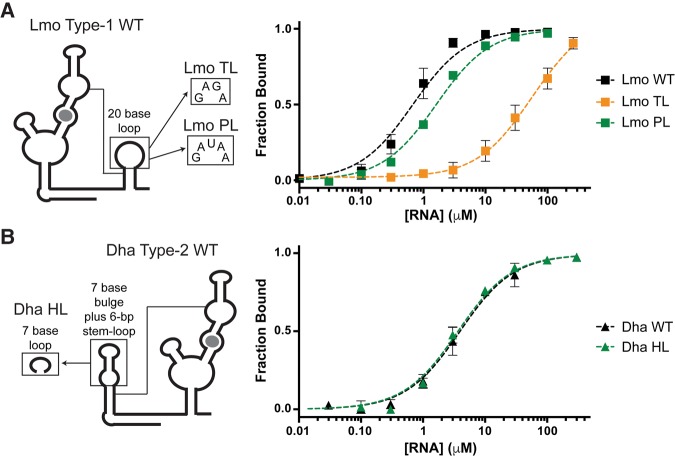
FIGURE 5.
Variable loops and helical extensions of singlet ghost aptamers can be deleted with minimal effect on glycine-binding affinity. (A,B) Diagrams and glycine-binding curves for singlet mutants with truncated or mutated loop regions.
Dha type-2 WT contains a 28-base extension of the ghost aptamer, which is predicted to form a 6-bp helical region with a bulge. As the helical extension was not conserved in other singlets, we deleted it, leaving a heptaloop (HL) with the sequence of the bulge (Table 3; Fig. 5B). Dha HL binds glycine as tightly as Dha WT. The Dha type-2 ghost-aptamer helical extension is, therefore, not involved in RNA folding or glycine binding.
A significant fraction of glycine riboswitches are singlets
We updated the consensus sequence for the glycine aptamer to be as general as possible (Supplemental Fig. S5) and searched the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) Database. Only sequences from completed genomes were considered for inclusion, in order to minimize false positives where aptamers could lie at the edge of contiguous sequence in a partly assembled genome. This search identified approximately 1000 unique glycine aptamers, which were then sorted into tandem or singlet riboswitches, where a tandem riboswitch was defined as two glycine aptamers that reside in the 5′-UTR of the same gene in an organism. The isolated aptamer domains were confirmed to be singlets by manual inspection. This process identified 322 glycine singlet riboswitches (50% of total).
CMfinder (Yao et al. 2006) indicates that there are conserved helices in the flanking sequences around every isolated aptamer domain. However, these helix sequences lack consistently conserved nucleotides that could definitively identify them as ghost aptamers. We speculate that the large number of representatives and the broader evolutionary diversity of host organisms in the full alignment may obscure any conserved sequences within the flanking helices. Because multiple helices often surround single aptamers, we were not able to sort the 322 singlets in the larger alignment into subtypes. Given the strict conservation of the A-rich bulge in the aptamer domain (Supplemental Fig. S5), the presence of flanking helices, and the known sequence flexibility of the helix portion of A-minor motifs (Doherty et al. 2001), the singlets in the larger alignment are likely to form tertiary interactions with peripheral helices.
We analyzed the genomic contexts of the isolated singlet aptamers and found that they are widely distributed, with examples identified in Gram-positive and Gram-negative bacteria (Fig. 6A). The singlet riboswitches are located in the 5′ untranslated regions (UTRs) of similar genes to tandem riboswitches (Fig. 6B). More than 50% regulate ORFs related to the glycine cleavage system. This suggests that these isolated aptamer domains are likely to be active riboswitches.
FIGURE 6.
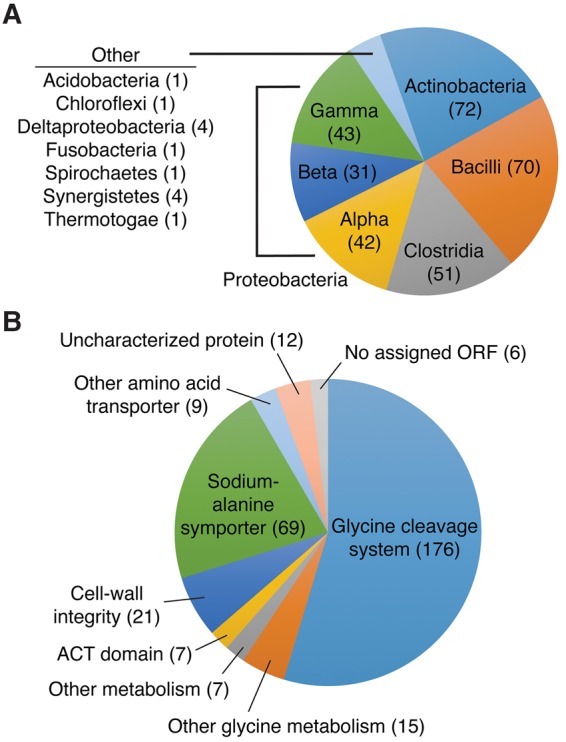
Distribution of singlet glycine riboswitches in the full alignment of 322 sequences. (A) Singlet glycine riboswitches are broadly distributed across bacterial phyla. (B) Singlet glycine riboswitches control genes associated with glycine metabolism, amino acid transport and metabolism, and cell-wall integrity.
DISCUSSION
In this study, we characterized the consensus motifs and in vitro ligand-binding activity of previously uncharacterized singlet glycine riboswitches. We identified 322 singlet glycine riboswitches, which represent a significant fraction of the glycine riboswitches in the database, singlet or tandem. Like the tandem riboswitches, the singlets are widely distributed, appear in similar genomic contexts, and are located in genes important for glycine homeostasis. Given the prevalence of singlet riboswitches in both Gram-positive and -negative bacteria, they are likely to be functional for gene control in both transcriptional (kinetic) and translational (thermodynamic) systems (Vitreschak et al. 2002; Barrick et al. 2004; Hollands et al. 2012).
Singlet riboswitches consist of a single glycine-aptamer domain flanked by a short stem–loop, here referred to as a “ghost aptamer.” The ghost-aptamer helical region maintains the length and sequence of the P1 stem of a standard glycine aptamer, while the loop varies greatly in length and sequence. Biochemical characterization of prototypical type-1 and type-2 singlets from L. monocytogenes and D. hafniense, respectively, demonstrated that both subtypes of singlet bind glycine with low micromolar affinities, comparable to tandem riboswitches. Mutational analysis of conserved binding and structural motifs within these singlets confirmed that the singlets contain a single canonical glycine-binding site and require the formation of tertiary A-minor interactions between the aptamer domain and the flanking stem–loop for ligand-binding activity.
Based on these observations, we propose a model for gene control by singlet glycine riboswitches (Fig. 7). It is informed by our model for tandem riboswitch function (Ruff and Strobel 2014), in which ligand binding in either aptamer stabilizes the tertiary interactions between the two aptamers, directly and indirectly stabilizing the P1 stem of aptamer-2. The formation of this stem competes with an alternative stem–loop, which hides or reveals the gene-control element (often an intrinsic terminator or the Shine-Dalgarno sequence) (Mandal et al. 2004; Ruff and Strobel 2014). According to this model, the conserved difference in P1-stem length and stability between the two aptamers plays a key role in stabilizing or scaffolding the formation of aptamer-2, either thermodynamically or kinetically (Ruff and Strobel 2014).
FIGURE 7.
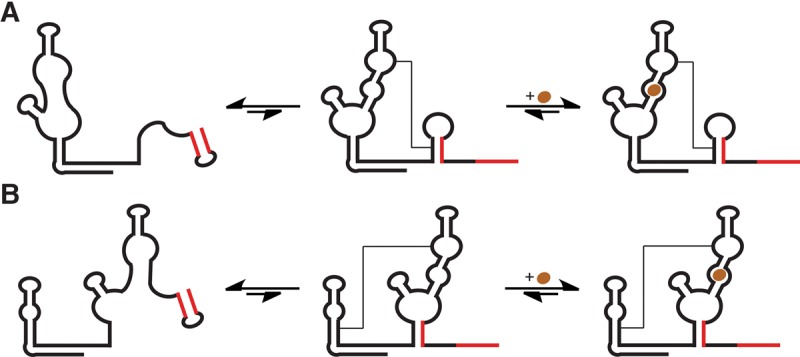
Proposed models for switch control for a type-1 singlet (A) or type-2 singlet (B). In each case, glycine binding stabilizes the overall tertiary structure, which destabilizes the alternative stem–loop responsible for gene control (red).
For a type-1 singlet, we propose that indirect stabilization of the ghost-aptamer stem plays a crucial role in gene control (Fig. 7A). Glycine binding in the aptamer domain promotes formation of the A-minor tertiary interaction, stabilizing the stem of the ghost-aptamer and disfavoring formation of the alternative stem–loop structure (shown in red). The aptamer domains of type-1 singlets have significantly more stable P1 stems than the aptamers of type-2. We propose that the P1 stem of this aptamer is preformed, similar to aptamer-1 in the tandem system (Sherman et al. 2012; Esquiaqui et al. 2014; Ruff and Strobel 2014; Cheng et al. 2015). Therefore, the energy of ligand binding is transferred, by way of the A-minor interaction, to the shorter, less stable stem of the ghost aptamer.
In contrast, for the type-2 singlet, we predict that ligand binding directly stabilizes the stem of the aptamer domain, disfavoring formation of the alternative stem–loop that serves as the switch for gene control (Fig. 7B). In this case, the ghost aptamer contains a longer, more stable stem–loop sequence that is likely preformed. This stabilizes or scaffolds the formation of the A-minor tertiary interaction, such that ligand binding stabilizes the short P1 stem of the aptamer domain.
Given the model above and the prevalence of singlet glycine riboswitches, we expect that singlets will be functional to control expression of their associated genes. At the same time, the conservation of two tandem glycine aptamers in the remaining ∼50% of cases indicates that some benefit is likely to be provided by the tandem system, such as more digital gene control, as originally proposed (Mandal et al. 2004). An examination of other riboswitch systems that bind two or more ligand molecules provides insight into potential benefits.
While the glycine riboswitch remains the only example of a tandem riboswitch wherein two homologous aptamer domains each bind a separate molecule of ligand and control a single expression platform, two other families of riboswitches have recently been shown to bind two molecules of ligand within a single aptamer domain. Both the tetrahydrofolate (THF) riboswitch (Trausch et al. 2011) and the cyclic-di-adenosine monophosphate (c-di-AMP) riboswitch (Gao and Serganov 2014; Ren and Patel 2014) were crystallized with two ligand molecules bound. In the case of the THF riboswitch, mutants that disrupt each ligand-binding site were identified and analyzed (Trausch and Batey 2014). In functional assays measuring transcriptional termination, only one of the two single-binding mutants responds to ligand. The authors propose that binding at the pseudoknot site is critical for gene control, while binding at the distal site could play a scaffolding role and/or allow a cooperative response to changes in THF concentration. In the case of the c-di-AMP riboswitch, ligand binding at the two sites appears to be cooperative (Meehan et al. 2016); although it is not yet clear if binding of both ligands is necessary for gene control.
There are multiple instances of tandem riboswitches where two or more complete riboswitches, each with an independent expression platform, control expression of a single open reading frame (ORF) (Sudarsan et al. 2006; Breaker 2012; Peselis et al. 2015; P McCown and R Breaker, unpubl.). The riboswitches can recognize the same ligand, as in the tandem thiamin pyrophosphate (TPP) riboswitches in Bacillus anthracis, where each riboswitch modulates an intrinsic terminator stem, leading to improved dose-response (Sudarsan et al. 2006; Welz and Breaker 2007; Breaker 2012). In Candidatus pelagibacter ubique, two different S-adenosylmethionine (SAM) riboswitch architectures control tandem expression platforms, the first transcriptional, the second translational (Poiata et al. 2009). The transcriptional riboswitch has been proposed to prevent wasteful transcription at very high SAM concentrations, whereas the translational riboswitch might allow a rapid increase in gene expression once ligand concentrations decrease (Poiata et al. 2009; Breaker 2012). Alternatively, tandem riboswitches can recognize different ligands, combining to form AND, NAND, or NOR logic gates. For example, the metE gene in Bacillus clausii is controlled by both SAM and adenosylcobalamin riboswitches (Sudarsan et al. 2006; Breaker 2012).
In the most similar case to glycine, glutamine riboswitches often contain two or even three aptamer sequences in tandem (Ames and Breaker 2011). Although no expression platform has been identified in these systems, the aptamers are separated by only a few nucleotides, and are unlikely to each control an individual expression platform. These tandem glutamine riboswitches do not bind cooperatively by in vitro ligand-binding assay (Ames and Breaker 2011), and no benefit of the tandem arrangement has been identified.
These other cases of tandem and dual-binding aptamers provide insight into the possible value of the tandem architecture for the glycine riboswitch. This study demonstrates that the previously characterized tandem glycine riboswitch does not provide improved ligand-binding affinity in vitro compared to singlets. Yet the prevalence of the tandem architecture implies that it provides some benefit. Perhaps the two aptamer domains are cooperative when controlling gene expression in vivo, providing a sharper, more digital response to changes in glycine concentration (Mandal et al. 2004). Multiple studies have shown that the isolated aptamer domains do not bind glycine cooperatively in vitro (Sherman et al. 2012; Baird and Ferré-D'Amaré 2013; Ruff and Strobel 2014), but perhaps, in the context of the full-length transcript, additional tertiary contacts with the expression platform affect the thermodynamics of ligand binding.
Alternatively, tandem riboswitches might be more effective for certain types of gene control or more versatile in accommodating different types of gene control, in the form of transcriptional versus translational or “ON” versus “OFF” switches. The glycine riboswitch class is one of only two classes that participate in both “ON” and “OFF” switches (the other being lysine) (Barrick and Breaker 2007). Perhaps the tandem architecture is more robust and therefore more easily transferrable from one context to another.
It is also interesting to speculate on the evolutionary history of the tandem and singlet architectures. The flanking “ghost-aptamer” sequences in our initial alignment were extremely highly conserved and almost certainly resulted from the deletion of the bulk of one aptamer from a tandem riboswitch. However, the tandem architecture itself likely resulted from the duplication of a progenitor singlet aptamer. Given the high prevalence of these riboswitches across bacterial phyla, perhaps this duplication and deletion process has occurred multiple times.
MATERIALS AND METHODS
DNA oligonucleotides and chemicals
DNA oligonucleotides were synthesized by the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. Overlapping oligonucleotides for the initial construction of the riboswitch templates were PAGE- or cartridge-purified. Primers were used without purification. Glycine and other chemicals were obtained from Sigma.
DNA constructs
The L. monocytogenes and S. mutans singlet templates were constructed by ligation of four overlapping oligonucleotides. The D. hafniense template was constructed by PCR of two long overlapping oligonucleotides. The final templates consisted of the T7 promoter sequence followed by the riboswitch DNA sequence in the pUC19 (NEB) plasmid. Mutant riboswitch constructs were made by PCR using corresponding primers.
In vitro transcription and denaturing purification
Plasmid DNA encoding the singlet glycine riboswitches was linearized by restriction digest and used as template for transcription by T7 RNA polymerase. RNAs were transcribed in 40 mM Tris-HCl (pH 7.5 at 23°C), 4 mM spermidine, 10 mM DTT, 55 mM MgCl2, 0.05% Triton X-100, and 4 mM of each 5′-nucleotide triphosphate (7 mM for GTP) for 2 h at 37°C.
The majority of RNAs were prepared with a denaturing protocol. They were purified by 6% polyacrylamide gel electrophoresis (PAGE), eluted into 0.3 M NaOAc (pH 5.2), precipitated with ethanol, and resuspended in TB buffer (90 mM Tris-borate at pH 8.3) containing 10 mM MgCl2 and 100 mM KCl. RNA transcripts were then buffer-exchanged four times and concentrated using Amicon Ultra centrifugal filters.
RNA concentrations were determined by UV absorbance at 260 nm. Absorption coefficients were determined by digestion with Nuclease P1, according to established protocols (Cavaluzzi and Borer 2004; Wilson et al. 2014). Briefly, ∼1 nmol of RNA was incubated at 50°C for 1 h with 1 unit of Nuclease P1 in 200 mM NaOAc, pH 5.3, with 5 mM EDTA and 10 mM Zn(OAc)2. Based on extinction coefficients for the individual nucleotides, the extinction coefficient of fully digested Lmo WT is 1.6 mol−1 cm−1 and that of the intact, folded RNA is 1.4 mol−1 cm−1. The extinction coefficients for fully digested and intact, folded Dha WT are 1.7 and 1.1 mol−1 cm−1, respectively.
Native FPLC purification of Dha WT
Dha WT was prepared using both the denaturing prep outlined above and a native preparation, as follows. In vitro-transcribed RNA was buffer exchanged several times into TB buffer containing 10 mM MgCl2 and 100 mM KCl using a 15-mL Macrosep Centrifugal Device (Pall Corporation). RNAs were purified by size-exclusion chromatography with a Superdex 200 column (GE Healthcare) on an NGC Chromatography System (Bio-Rad) with TB running buffer containing 10 mM MgCl2 and 100 mM KCl. Monomer fractions were combined, concentrated using Amicon Ultra centrifugal filters, and quantified as discussed above. Analytical FPLC under the same conditions confirmed that the monomer Dha WT is stable over the time frame of the dialysis experiments.
Equilibrium dialysis assay
RNA transcripts were combined with trace 14C-labeled glycine in TB buffer containing 10 mM MgCl2 and 100 mM KCl. Samples were heated to 60°C then allowed to slow cool to ∼30°C over an hour. The RNA/glycine mixture was equilibrated overnight at 23°C across from an equal volume of buffer in a 5000 MW cut-off Dispo Equilibrium Dialyzer from Harvard Apparatus. The amount of 14C-labeled glycine on each side of the dialyzer was determined by scintillation counting in Ultima Gold on a PerkinElmer Tri-Carb 2910TR scintillation counter. The fraction bound was determined for each sample ([counts on RNA side − counts on buffer side]/counts on RNA side). The Kd value for glycine binding was determined by plotting the fraction-bound value versus the concentration of RNA and fitting to a standard equation for one-site binding, using Prism to perform a least squares regression:
| (1) |
where Y is the fraction bound, X is the concentration of RNA, and NS is a constant term for nonspecific binding.
For mutants that failed to saturate at the RNA concentrations tested, Bmax values were fixed at 0.99. Errors were calculated based on at least four replicates from at least two independent preparations of RNA.
Equilibrium dialysis equivalents assay
RNA transcripts were combined with a threefold excess of cold glycine as well as trace 14C-labeled glycine. The RNA was refolded and equilibrated with buffer in a 5000 MW cut-off equilibrium dialysis cassette, as discussed above. The amount of 14C-labeled glycine on each side of the dialyzer was determined by scintillation counting, and the equivalents of bound glycine were determined ([counts on RNA side – counts on buffer side] × 3/[counts on RNA side + counts on buffer side]). Errors are standard deviations calculated based on at least six replicates from at least two independent preparations of RNA.
Native equilibrium dialysis equivalents assay
Natively prepared Dha WT was combined with a threefold excess of cold glycine as well as trace 14C-labeled glycine. The RNA was not refolded. Instead, it was equilibrated overnight with buffer in a 5000 MW cut-off equilibrium dialysis cassette. The equivalents of bound glycine were determined as discussed above.
Multi-angle light scattering
Lmo RNA was transcribed and buffer-exchanged as discussed above. Glycine was added to a final concentration of 5 mM, and the RNA was heated to 60°C then allowed to slow cool to ∼30°C over an hour before further incubation at room temperature for at least 30 min. Thirty-micromolar samples were filtered and analyzed without further dilution. Three hundred-micromolar samples were diluted 10-fold and filtered immediately before analysis.
Natively prepared Dha WT was combined with a threefold excess of cold glycine. The RNA was not refolded. Instead, it was equilibrated for 5 h at room temperature, then diluted and filtered immediately before analysis.
RNAs were separated by size-exclusion on a Superdex 200 column (GE Healthcare) with TB running buffer containing 10 mM MgCl2 and 100 mM KCl. Light-scattering data were collected on a DAWN HELEOS (Wyatt Technology) coupled to an OPTILAB rEX (Wyatt Technology) interferometric refractometer. Multi-angle light scattering (660 nm), absorbance (280 nm), and the refractive index were monitored after elution. Molar masses were determined by comparison of light scatter to the monomeric fraction of a BSA control (Sigma-Aldrich).
Analysis of multimeric state by size-exclusion FPLC
RNA transcripts were buffer exchanged into TB running buffer containing 10 mM MgCl2 and 100 mM KCl, as discussed above. Five-millimolar glycine was added where indicated, then the RNAs were heated to 60°C and allowed to slow cool to ∼30°C over an hour before further incubation at room temperature for at least 30 min. Samples were diluted and filtered immediately before analysis.
The oligomeric, dimeric, and monomeric fractions of the RNAs were separated by size-exclusion chromatography with a Superdex 200 column (GE Healthcare) on an NGC Chromatography System (Bio-Rad) with TB running buffer containing 10 mM MgCl2 and 100 mM KCl. Peaks in the UV absorbance (255 nM) were integrated using ChromLab software (Bio-Rad), with peak cutoffs manually adjusted to be consistent between consecutive runs. Errors are standard deviations calculated from two or three replicates from two independent preparations of RNA.
Bioinformatics searches and alignment generation
Bioinformatics searches for all glycine riboswitches, including possible variant glycine riboswitches, were conducted as previously described (McCown et al. 2011, 2014). Using INFERNAL 1.1, searches were conducted on the National Center for Biotechnology Information (NCBI) Reference Sequence (RefSeq) Database release 56, with the highest E-value for possible variants set to 1000. Only sequences from completed genomes in this data set were considered for inclusion, so as to minimize erroneously reported singlets. Once a list of all glycine riboswitches was determined from this data set, the consensus structure markup was changed to remove possible discrimination between type-1 and type-2 aptamers (Mandal et al. 2004). The resulting Stockholm file was then used as a template for an additional search through the same NCBI RefSeq database as mentioned above. False-positive results were eliminated based on overlap with a known ORF or where the sequence lacked key nucleotides that are necessary to bind glycine (Huang et al. 2010; Butler et al. 2011). As the generalized Stockholm file contains only one aptamer, the resulting riboswitch results were then determined to be either singlet arrangements or tandem arrangements, with the definition of tandem as being within the same 5′-UTR with another glycine riboswitch within the same organism. Each singlet riboswitch was verified by examining the entire 5′-UTR for any additional glycine aptamers. Riboswitch consensus diagrams were generated with the R2R program (Weinberg and Breaker 2011).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeff Barrick, Ethan Butler, Zasha Weinberg, David Hiller, Cambria Alpha-Cobb, and Kathryn Smith for helpful discussions. We thank David Hiller for critical comments on the manuscript, William Eliason of the Yale Center for Structural Biology Core for MALS analysis, and Patricia Gordon for transcription reagents and general laboratory support. We thank Nick Carriero and Rob Bjornson for assisting in our use of the Yale Life Sciences High Performance Computing Center (National Institutes of Health [NIH] grant RR19895-02). This work was supported by National Institutes of Health grant GM022778. P.J.M. was supported in part by National Institutes of Health grant T32GM007499. R.R.B. is a Howard Hughes Medical Institute investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.057935.116.
REFERENCES
- Ames TD, Breaker RR. 2011. Bacterial aptamers that selectively bind glutamine. RNA Biol 8: 82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NJ, Ferré-D'Amaré AR. 2013. Modulation of quaternary structure and enhancement of ligand binding by the K-turn of tandem glycine riboswitches. RNA 19: 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 43: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR. 2012. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol 4: a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. 2011. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol 18: 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaluzzi MJ, Borer PN. 2004. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res 32: e13–e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Chou F-C, Kladwang W, Tian S, Cordero P, Das R. 2015. Consistent global structures of complex RNA states through multidimensional chemical mapping. Elife 4: e07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty EA, Batey RT, Masquida B, Doudna JA. 2001. A universal mode of helix packing in RNA. Nat Struct Mol Biol 8: 339–343. [DOI] [PubMed] [Google Scholar]
- Erion TV, Strobel SA. 2011. Identification of a tertiary interaction important for cooperative ligand binding by the glycine riboswitch. RNA 17: 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquiaqui JM, Sherman EM, Ionescu SA, Ye J-D, Fanucci GE. 2014. Characterizing the dynamics of the leader–linker interaction in the glycine riboswitch with site-directed spin labeling. Biochemistry 53: 3526–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao A, Serganov A. 2014. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat Chem Biol 10: 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. 2012. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci 109: 5376–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Serganov A, Patel DJ. 2010. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell 40: 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazanov MD, Vitreschak AG, Gelfand MS. 2007. Abundance and functional diversity of riboswitches in microbial communities. BMC Genomics 8: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kladwang W, Chou F-C, Das R. 2012. Automated RNA structure prediction uncovers a kink-turn linker in double glycine riboswitches. J Am Chem Soc 134: 1404–1407. [DOI] [PubMed] [Google Scholar]
- Kwon M, Strobel SA. 2008. Chemical basis of glycine riboswitch cooperativity. RNA 14: 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J, Das R, Chu VB, Kudaravalli M, Boyd N, Herschlag D, Doniach S. 2007. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol 365: 1393–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306: 275–279. [DOI] [PubMed] [Google Scholar]
- McCown PJ, Roth A, Breaker RR. 2011. An expanded collection and refined consensus model of glmS ribozymes. RNA 17: 728–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCown PJ, Liang JJ, Weinberg Z, Breaker RR. 2014. Structural, functional, and taxonomic diversity of three preQ1 riboswitch classes. Chem Biol 21: 880–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan RE, Torgerson CD, Gaffney BL, Jones RA, Strobel SA. 2016. Nuclease-resistant c-di-AMP derivatives that differentially recognize RNA and protein receptors. Biochemistry 55: 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis A, Gao A, Serganov A. 2015. Cooperativity, allostery and synergism in ligand binding to riboswitches. Biochimie 117: 100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poiata E, Meyer MM, Ames TD, Breaker RR. 2009. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 15: 2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A, Patel DJ. 2014. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat Chem Biol 10: 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff KM, Strobel SA. 2014. Ligand binding by the tandem glycine riboswitch depends on aptamer dimerization but not double ligand occupancy. RNA 20: 1775–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman EM, Esquiaqui J, Elsayed G, Ye J-D. 2012. An energetically beneficial leader-linker interaction abolishes ligand-binding cooperativity in glycine riboswitches. RNA 18: 496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR. 2006. Tandem riboswitch architectures exhibit complex gene control functions. Science 314: 300–304. [DOI] [PubMed] [Google Scholar]
- Tezuka T, Ohnishi Y. 2014. Two glycine riboswitches activate the glycine cleavage system essential for glycine detoxification in Streptomyces griseus. J Bacteriol 196: 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Batey RT. 2014. A disconnect between high-affinity binding and efficient regulation by antifolates and purines in the tetrahydrofolate riboswitch. Chem Biol 21: 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trausch JJ, Ceres P, Reyes FE, Batey RT. 2011. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure 19: 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 30: 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Breaker RR. 2011. R2R—software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics 12: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz R, Breaker RR. 2007. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 13: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SC, Cohen DT, Wang XC, Hammond MC. 2014. A neutral pH thermal hydrolysis method for quantification of structured RNAs. RNA 20: 1153–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Weinberg Z, Ruzzo WL. 2006. CMfinder—a covariance model based RNA motif finding algorithm. Bioinformatics 22: 445–452. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jones CP, Ferré-D'Amaré AR. 2014. Global analysis of riboswitches by small-angle X-ray scattering and calorimetry. Biochim Biophys Acta 1839: 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.