Summary
The importance of the nitrate () transporter for yield and nitrogen‐use efficiency (NUE) in rice was previously demonstrated using map‐based cloning. In this study, we enhanced the expression of the OsNRT2.1 gene, which encodes a high‐affinity transporter, using a ubiquitin (Ubi) promoter and the ‐inducible promoter of the OsNAR2.1 gene to drive OsNRT2.1 expression in transgenic rice plants. Transgenic lines expressing pUbi:OsNRT2.1 or pOsNAR2.1:OsNRT2.1 constructs exhibited the increased total biomass including yields of approximately 21% and 38% compared with wild‐type (WT) plants. The agricultural NUE (ANUE) of the pUbi:OsNRT2.1 lines decreased to 83% of that of the WT plants, while the ANUE of the pOsNAR2.1:OsNRT2.1 lines increased to 128% of that of the WT plants. The dry matter transfer into grain decreased by 68% in the pUbi:OsNRT2.1 lines and increased by 46% in the pOsNAR2.1:OsNRT2.1 lines relative to the WT. The expression of OsNRT2.1 in shoot and grain showed that Ubi enhanced OsNRT2.1 expression by 7.5‐fold averagely and OsNAR2.1 promoters increased by about 80% higher than the WT. Interestingly, we found that the OsNAR2.1 was expressed higher in all the organs of pUbi:OsNRT2.1 lines; however, for pOsNAR2.1:OsNRT2.1 lines, OsNAR2.1 expression was only increased in root, leaf sheaths and internodes. We show that increased expression of OsNRT2.1, especially driven by OsNAR2.1 promoter, can improve the yield and NUE in rice.
Keywords: OsNAR2.1 promoter, Oryza sativa, OsNRT2.1, agronomic nitrogen‐use efficiency
Introduction
Rice (Oryza sativa L.) is not only a major staple food crop for a large part of the world population but also an important model monocot plant species for research because of its small genome size and the availability of the complete rice genome sequence (Feng et al., 2002; Sasaki et al., 2002). Nitrogen (N) nutrition affects all levels of plant function from metabolism to resource allocation, growth and development (Crawford, 1995; Scheible et al., 1997, 2004; Stitt, 1999). The most abundant source for N acquisition by plant roots is nitrate (), which is present in naturally aerobic soils due to intensive nitrification from applied organic and fertilizer N. In contrast, ammonium () is the main form of available N in flooded paddy soils due to the anaerobic soil conditions (Sasakawa and Yamamoto, 1978).
serves as a nutrient and as a signal that induces changes in the growth and gene expression (Coruzzi and Bush, 2001; Coruzzi and Zhou, 2001; Crawford and Forde, 2002; Crawford and Glass, 1998; Kirk and Kronzucker, 2005; Kronzucker et al., 2000; Wang et al., 2000; Zhang and Forde, 2000). Two different uptake systems in plants, the high‐ and low‐affinity uptake systems designated as HATS and LATS, respectively, are regulated by supply and enable plants to cope with high or low concentrations in soils (Fan et al., 2005).
Some high‐affinity transporters belonging to the NRT2 family have been shown to require a partner protein, NAR2, for their function (Xu et al., 2012). Quesada et al. (1994) identified the CrNar2 gene, which encodes a small protein of approximately 200 amino acid residues and which has no known transport activity, but is required for complementation of transport in Chlamydomonas reinhardtii mutants defective in uptake. In Arabidopsis, Okamoto et al. (2006) showed that both constitutive and ‐inducible HATS, but not LATS, depended on the expression of the NAR2‐type gene, for example Arabidopsis AtNRT3.1. Orsel et al. (2006) used yeast split‐ubiquitin and oocyte expression systems to show that AtNAR2.1 (AtNRT3.1) and AtNRT2.1 interacted to produce a functional HATS. Yong et al. (2010) showed that the NRT2.1 and NAR2.1 polypeptides interact directly at the plasma membrane to constitute an oligomer that may act as the functional unit for high‐affinity influx in Arabidopsis roots. In rice, the OsNRT2.1, OsNRT2.2 and OsNRT2.3a gene products were similarly shown to require the protein encoded by OsNAR2.1 for uptake (Feng et al., 2011; Liu et al., 2014; Yan et al., 2011), and their interaction at the protein level was demonstrated using a yeast two‐hybrid assay and by Western blotting (Liu et al., 2014; Yan et al., 2011).
Rice seedling growth was improved slightly by increased OsNRT2.1 expression, but N uptake remained unaffected (Katayama et al., 2009), probably due to the absence of the interaction with OsNAR2.1, which is required for the functional transport (Feng et al., 2011; Yan et al., 2011).
In this study, we transformed the open reading frame (ORF) of the OsNRT2.1 gene into rice with the expression driven by the OsNAR2.1 promoter to modify the coexpression of the OsNRT2.1 and OsNAR2.1 genes in rice plants and to investigate the biological function of their coexpression in vivo. Transgenic lines expressing the OsNRT2.1 gene under the control of the OsNAR2.1 promoter exhibited greatly increased the growth, biomass and yield compared with transgenic lines expressing OsNRT2.1 under a ubiquitin promoter. We analysed OsNRT2.1 and OsNAR2.1 expression patterns during the whole‐plant growth and show that modification of the ratio of OsNRT2.1 to OsNAR2.1 expression in stems altered the rice growth and agricultural N‐use efficiency (ANUE).
Results
Generation of transgenic rice plants expressing pUbi:OsNRT2.1 and pOsNAR2.1:OsNRT2.1 constructs and field analysis of traits
The ubiquitin promoter (pUbi) has been used as a strong promoter in a variety of applications in gene transfer studies and was shown to drive gene expression most actively in rapidly dividing cells (Cornejo et al., 1993). Overexpression of just the OsNRT2.1 gene in rice was previously shown to not increase uptake (Katayama et al., 2009).
We introduced pUbi:OsNRT2.1 (Figure S1a) and pOsNAR2.1:OsNRT2.1 (Figure S1b) expression constructs into Wuyunjing 7 (WYJ7), a rice cultivar that produces high yields in Jiangsu Province, using Agrobacterium tumefaciens‐mediated transformation. We generated 23 lines exhibiting increased OsNRT2.1 expression, including 12 pUbi:OsNRT2.1 lines and 11 pOsNAR2.1:OsNRT2.1 lines (Figure S2).
We analysed the grain yield and biomass of transgenic lines in the T0 and T1 generations. Relative to the wild‐type (WT) plants, the biomass, including the grain yield, of the 12 pUbi:OsNRT2.1 lines increased by approximately 21.8% (Figure S2e) and 20.9% (Figure S3a) in T0 and T1 plants, respectively, but the grain yield decreased approximately 18.4% (Figure S2c) and 16.6% (Figure S3a) in T0 and T1 plants, respectively. Relative to the WT, the biomass, including the grain yield, of the 11 pOsNAR2.1:OsNRT2.1 lines increased by average values of 32.2% (Figure S2f) and 27.1% (Figure S3b) in T0 and T1 plants, respectively, and the grain yield increased by average values of 30.7% (Figure S2d) and 28.1% (Figure S3b) in T0 and T1 plants, respectively. Based on the Southern blot analysis of T1 plants (Figure S4) and RNA expression data for the T0 generation (Figure S2a,b), we selected three independent pUbi:OsNRT2.1 T1 lines OE1–2, OE2–5 and OE3–4 [renamed as OE1, OE2 and OE3 (Figure 1a)] and three independent pOsNAR2.1:OsNRT2.1 T1 lines O6–4, O7–6 and O8–3 [renamed as O6, O7 and O8 (Figure 1b)].
Figure 1.
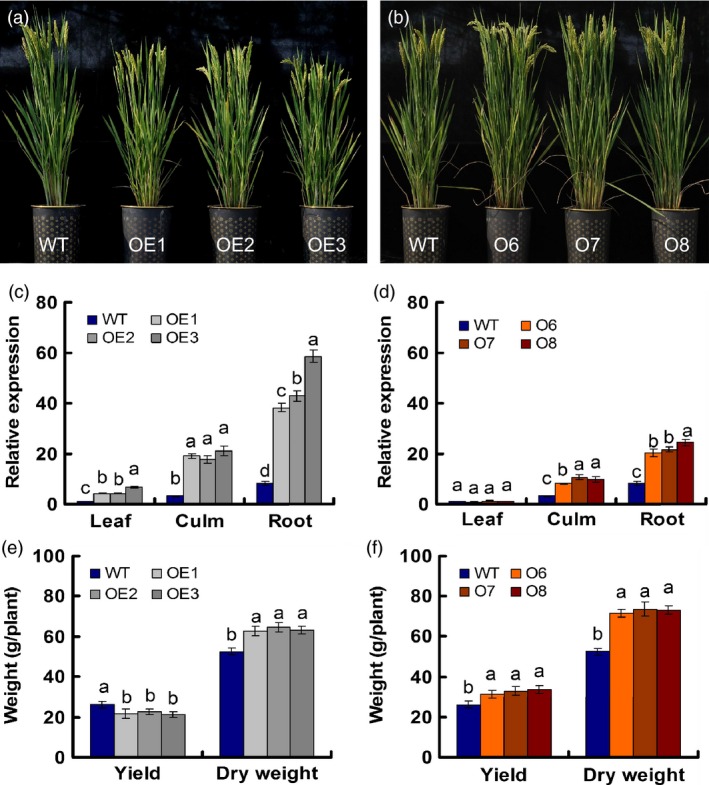
Characterization of transgenic lines. (a) Gross morphology of pUbi:OsNRT2.1 transgenic lines (OE1, OE2 and OE3) and the wild‐type (WT). (b) Gross morphology of pOsNAR2.1:OsNRT2.1 transgenic lines (O6, O7 and O8) and the WT. (c, d) Real‐time quantitative RT‐PCR analysis of endogenous OsNRT2.1 expression in various transgenic lines and WT plants. (c) pUbi:OsNRT2.1 transgenic lines (OE1, OE2 and OE3) and the WT, and (d) pOsNAR2.1:OsNRT2.1 transgenic lines (O6, O7 and O8) and the WT. RNA was extracted from leaf blade I, culm and root. (e, f) Grain yield and dry weight per plant for transgenic and WT plants grown in the field. Dry weight mean values are for all aboveground biomass, including the grain yield. (e) pUbi:OsNRT2.1 transgenic lines and WT. (f) pOsNAR2.1:OsNRT2.1 transgenic lines and WT. Statistical analysis was performed on data derived from the T3 generation. Error bars: SE (n = 3). Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA).
Agricultural traits of these six lines were investigated in the field in the T1 through T4 generations, with a particular focus on the T3 generation. OsNRT2.1 expression in roots was enhanced four‐ to sevenfold in the OE1, OE2 and OE3 lines but only 2.5‐ to threefold in the O6, O7 and O8 lines relative to the WT. In culms, OsNRT2.1 expression was increased approximately sixfold in the OE lines and approximately threefold in the O lines. In leaf blades, however, only the OE lines exhibited increased OsNRT2.1 expression (four‐ to sevenfold) compared with the WT, and no change in the expression was observed in the O lines (Figure 1c,d). The field data showed that both the OE and O lines exhibited increased growth and biomass, but only the O lines produced higher yields than the WT (Figure 1e,f).
Based on the agricultural traits of the T1–T4 generation plants in the field, the total aboveground biomass including the grain yield increased by 21% for the pUbi:OsNRT2.1 lines and by 38% for the pOsNAR2.1:OsNRT2.1 lines, while the biomass without grain yield increased by 190% for the pUbi:OsNRT2.1 lines and by 160% for the pOsNAR2.1:OsNRT2.1 lines. The grain yields of the pUbi:OsNRT2.1 lines decreased over the three successive generations (Table 1), but the yields of the pOsNAR2.1:OsNRT2.1 lines increased significantly from the T1 to T3 generation (Table 1). The yields of the O lines were enhanced by approximately 33% in T1 plants grown at Ledong and by 34%–42% in the T2 and T3 generations grown at Nanjing relative to the WT, while the OE lines exhibited lower yields than the WT by approximately 17% in all three generations (Table 1). We also analysed the yield and the biomass of the WT and T4 generation transgenic plants at Nanjing under low (180 kg N/ha) and normal N (300 kg N/ha) supplies. At the level of 180 kg N/ha, compared with WT, the yield of OE lines was reduced by 17%, and the biomass increased by 14%, while the yield and biomass of O lines were increased by 25% and 27% (Figure S5a), respectively. At the level of 300 kg N/ha, the yield of OE lines was reduced by 16%, and the biomass increased by 12%, as for O lines the yield and biomass were increased by 21% and 22%, respectively, compared with WT (Figure S5b).
Table 1.
Comparison of the grain yield, dry weight and agronomic nitrogen‐use efficiency (ANUE) between the wild‐type (WT) and transgenic lines in the T1–T3 generations
| WT | pUBi:OsNRT2.1 | pOsNAR2.1:OsNRT2.1 | |||||
|---|---|---|---|---|---|---|---|
| OE1 | OE2 | OE3 | O6 | O7 | O8 | ||
| Grain yield (kg/m2) | |||||||
| T1 | 0.52 b | 0.42 c | 0.44 c | 0.43 c | 0.69 a | 0.69 a | 0.71 a |
| T2 | 0.66 b | 0.54 c | 0.56 c | 0.54 c | 0.89 a | 0.91 a | 0.93 a |
| T3 | 0.70 b | 0.58 c | 0.60 c | 0.57 c | 0.94 a | 0.98 a | 1.00 a |
| Dry weight (kg/m2) | |||||||
| T1 | 1.05 c | 1.31 b | 1.29 b | 1.31 b | 1.43 a | 1.45 a | 1.47 a |
| T2 | 1.27 c | 1.55 b | 1.61 b | 1.58 b | 1.77 a | 1.83 a | 1.77 a |
| T3 | 1.40 c | 1.67 b | 1.73 b | 1.69 b | 1.91 a | 1.96 a | 1.95 a |
| ANUE (g/g) | |||||||
| T1 | 15.48 b | 12.43 c | 12.94 c | 12.56 c | 19.64 a | 19.63 a | 19.86 a |
| T2 | 20.28 b | 16.46 c | 17.02 c | 16.25 c | 26.41 a | 26.71 a | 27.50 a |
| T3 | 21.33 b | 17.42 c | 18.41 c | 17.01 c | 26.17 a | 27.86 a | 28.12 a |
Dry weight mean values are for all aboveground biomass, including the grain yield. For each mean, n = 3. Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA).
The total tiller number per plant in the T3 generation at the harvest stage increased 27.1% on average for both pOsNAR2.1:OsNRT2.1 and pUbi:OsNRT2.1 transgenic plants relative to the WT with no difference between the transgenic lines (Table 2); however, the grain number per panicle differed significantly between the OE and O lines (Table 2). The grain number per panicle increased approximately 15% in the O lines; the panicle length increased in the O lines approximately 12%, and the seed setting rate increased in the O lines by 14% relative to the WT (Table 2). The grain yields of the O lines increased by 24.2% relative to the WT (Table 2).
Table 2.
Comparison of agronomic traits between the wild‐type (WT) and transgenic lines
| Genotype | WT | pUBi:OsNRT2.1 | pOsNAR2.1:OsNRT2.1 | ||||
|---|---|---|---|---|---|---|---|
| OE1 | OE2 | OE3 | O6 | O7 | O8 | ||
| Plant height (cm) | 83.21 b | 80.18 c | 79.25 c | 76.25 d | 87.27 a | 86.85 a | 88.69 a |
| Total tiller number per plant | 20.26 b | 25.57 a | 23.24 a | 24.81 a | 25.83 a | 26.84 a | 24.41 a |
| Panicle length (cm) | 13.19 b | 12.48 c | 11.48 c | 11.12 c | 14.40 a | 14.13 a | 14.48 a |
| Grain weight (g/panicle) | 2.22 d | 1.16 e | 0.96 f | 1.23 e | 3.87 a | 2.74 c | 3.01 b,c |
| Seed setting rate (%) | 70.45 b | 59.79 c | 57.82 c | 61.94 c | 80.46 a | 75.92 a | 78.95 a |
| Grain number per panicle | 132.58 b | 105.67 c | 97.25 c | 101.61 c | 154.50 a | 166.25 a | 149.75 a |
| 1000‐grain weight (g) | 25.24 a | 24.89 a | 24.39 a | 24.45 a | 25.28 a | 25.67 a | 25.89 a |
| Grain yield (g/plant) | 26.21 b | 21.61 c | 22.63 c | 21.19 c | 31.17 a | 32.81 a | 33.64 a |
Statistical analysis was performed on data derived from the T3 generation. Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA, n = 3).
Nitrogen‐use efficiency of transgenic lines
Because the biomass and yields increased in the pOsNAR2.1:OsNRT2.1 transgenic plants, we also analysed ANUE in T1–T4 generations of transgenic plants, N recovery efficiency (NRE), physiological N‐use efficiency (PNUE) and N harvest index (NHI) traits at the harvest stage in T3 generation transgenic lines to determine whether N use was altered in these plants, as modified the calculation method of the reference in Zhang et al. (2009). The ANUE of the O lines was enhanced by approximately 33% in T1 plants grown at Ledong and by 34%–42% in the T2 and T3 generations grown at Nanjing relative to the WT, while the OE lines exhibited lower ANUE than the WT by approximately 17% in all three generations (Table 1). In T4 plants at Nanjing, at the level of 180 kg N/ha, compared with WT, the ANUE of OE lines was reduced by 22% and the ANUE of O lines was increased by 33%, and at the level of 300 kg N/ha, the ANUE of OE lines was reduced by 17% and the ANUE of O lines was increased by 28% (Figure S5c). In the OE lines, the NRE increased to approximately 115% of the WT, and the PNUE and NHI were reduced to approximately 71% of the WT values. In the O lines, the ANUE increased to approximately 128% of the WT, the NRE increased to approximately 136% of the WT, and the PNUE and NHI were not significantly different from WT values (Table 4).
Table 4.
Comparison of N‐use efficiency, dry matter transport efficiency and N transport efficiency between the wild‐type (WT) and transgenic rice lines
| WT | pUBi:OsNRT2.1 | pOsNAR2.1:OsNRT2.1 | |||||
|---|---|---|---|---|---|---|---|
| OE1 | OE2 | OE3 | O6 | O7 | O8 | ||
| N recovery efficiency (%) | 39.06 c | 44.59 b | 45.40 b | 45.64 b | 52.13 a | 53.29 a | 53.68 a |
| Physiological N‐use efficiency (g/g) | 54.55 a | 39.96 b | 40.56 b | 37.26 b | 50.10 a | 51.49 a | 52.40 a |
| N harvest index (%) | 59.49 a | 43.52 b | 42.39 b | 41.31 b | 61.98 a | 62.68 a | 61.56 a |
| Dry matter transfer (g/m2) | 198.95 c | 72.25 d | 51.03 e | 67.74 d | 301.22 a | 278.87 b | 293.48 a,b |
| Dry matter transfer efficiency (%) | 22.10 a | 6.32 b | 4.45 c | 5.95 b | 23.23 a | 21.22 a | 21.78 a |
| Contribution of preanthesis assimilates to grain yield (%) | 28.45 a | 12.53 b | 8.45 c | 11.98 b | 30.10 a | 28.39 a | 29.22 a |
| Harvest index (%) | 49.93 a | 34.46 b | 34.96 b | 33.47 b | 49.20 a | 50.34 a | 51.46 a |
| Postanthesis N uptake (g/m2) | 2.64 c | 2.66 c | 2.83 c | 2.84 c | 4.45 b | 5.03 a | 5.40 a |
| N translocation (g/m2) | 6.91 b | 5.91 c | 5.84 c | 5.70 c | 8.24 a | 8.28 a | 7.93 a |
| N translocation efficiency (%) | 49.45 a | 34.69 b | 33.14 b | 31.97 b | 51.42 a | 51.10 a | 48.42 a |
| Contribution of preanthesis N to grain N accumulation (%) | 72.34 a | 68.95 a | 68.36 a | 69.76 a | 61.93 b | 62.21 b | 58.62 b |
Statistical analysis was performed on data derived from the T3 generation. Methods of calculations in Table S4. For each mean, n = 3. Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA).
We sampled shoot tissues at the anthesis stage (60 days after transplanting) and the mature stage (90 days after transplanting) to determine the total N content. At the anthesis stage, total N was concentrated mainly in the culm with no difference between the OE and O lines, but with an increase of approximately 27% relative to the WT. In leaves, the total N content was the same in the O and WT lines, but was approximately 33% higher in the OE lines. The total N content in the grain was the same in all lines (Figure 2a). At the mature stage, total N was concentrated mainly in the grain, with the N content decreased by approximately 10% in the OE lines and increased by approximately 38% in the O lines relative to the WT (Figure 2b).
Figure 2.
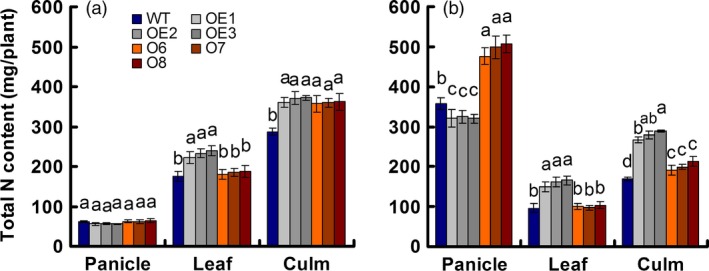
N content in various parts of the wild‐type (WT) and transgenic plants at two growth stages. (a) Sixty days after transplant, anthesis stage. (b) Ninety days after transplant, maturity stage. Error bars: SE (n = 3). Statistical analysis was performed on data derived from the T3 generation. Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA).
Translocation of dry matter and N in transgenic lines
We investigated the dry matter and N translocation (NT) in rice plants by determining dry matter at anthesis (DMA), dry matter at maturity (DMM), total N accumulation at anthesis (TNAA) and total N accumulation at maturity (TNAM). For the OE lines, the DMA, the DMM, the TNAA and the TNAM increased by approximately 27%, 21%, 25% and 21%, respectively, relative to the WT. For the O lines, the DMA, the DMM, the TNAA and the TNAM increased by approximately 46%, 38%, 15% and 27%, respectively, relative to the WT (Table 3).
Table 3.
Comparison of dry matter accumulation and N content between the wild‐type (WT) and transgenic lines
| Dry matter and nitrogen components | WT | pUBi:OsNRT2.1 | pOsNAR2.1:OsNRT2.1 | ||||
|---|---|---|---|---|---|---|---|
| OE1 | OE2 | OE3 | O6 | O7 | O8 | ||
| Dry matter at anthesis (kg/m2) | 0.90 c | 1.14 b | 1.15 b | 1.14 b | 1.30 a | 1.31 a | 1.35 a |
| Dry matter at maturity (kg/m2) | 1.40 c | 1.67 b | 1.78 b | 1.69 b | 1.91 a | 1.96 a | 1.95 a |
| Total nitrogen accumulation at anthesis (g/m2) | 13.98 c | 17.02 a | 17.62 a | 17.83 a | 16.02 b | 16.20 b | 16.38 b |
| Total nitrogen accumulation at maturity (g/m2) | 16.62 b | 19.68 a | 20.45 a | 20.67 a | 20.47 a | 21.22 a | 21.98 a |
| Grain nitrogen accumulation at maturity (g/m2) | 9.56 b | 8.56 c | 8.67 c | 8.54 c | 12.69 a | 13.30 a | 13.53 a |
Statistical analysis was performed on data derived from the T3 generation. For each mean, n = 3. Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA).
We also investigated the dry matter translocation (DMT), the DMT efficiency (DMTE), the contribution of preanthesis assimilates to grain yield (CPAY) and the harvest index (HI), based on the calculation method of the reference in Ntanos and Koutroubas (2002). For the OE lines, the DMT, DMTE, CPAY and HI decreased by approximately 68%, 75%, 61% and 31%, respectively, relative to the WT. For the O lines, the DMT increased by approximately 46%, while the DMTE, CPAY and HI did not differ between the O lines and the WT (Table 4).
We investigated postanthesis N uptake (PANU), NT, N translocation efficiency (NTE) and the contribution of preanthesis N to grain N accumulation (CPNGN), as modified the calculation method of the reference in Ntanos and Koutroubas (2002) and Zhang et al. (2009). The PANU and CPNGN did not differ between the OE lines and the WT, but the NT and the NTE decreased by approximately 16% and 32%, respectively, in the OE lines relative to the WT. The NTE did not differ between the O lines and the WT, while the PANU and NT increased by approximately 87% and 18%, respectively, and the CPNGN decreased by approximately 16% in the O lines relative to the WT (Table 4).
Expression patterns of OsNRT2.1 and OsNAR2.1 in different organs of the WT and transgenic lines
Rice was previously shown to have a two‐component uptake system consisting of OsNRT2.1 and OsNAR2.1, similar to the system in Arabidopsis (Feng et al., 2011; Liu et al., 2014; Yan et al., 2011). We analysed the OsNRT2.1 and OsNAR2.1 expression patterns in the WT and transgenic lines during the filling stage. The detail about RNA samples was described in Figure S6 and Experimental procedures. The OsNRT2.1 expression pattern in the WT showed that OsNRT2.1 gene expressed most in root, secondly in leaf sheaths, thirdly in leaf blades and internodes and lest in grain including seed, palea and lemma (Table S5, Figure 3a). As for OsNAR2.1, it was expressed also most in root, secondly in leaf sheaths, thirdly in internodes and lest in grain and leaf blades (Table S5, Figure 3b). The coexpression pattern of OsNRT2.1 and OsNAR2.1 happened in root, leaf sheaths, internodes and grain but not in leaf blades (Table S5, Figure S7).
Figure 3.
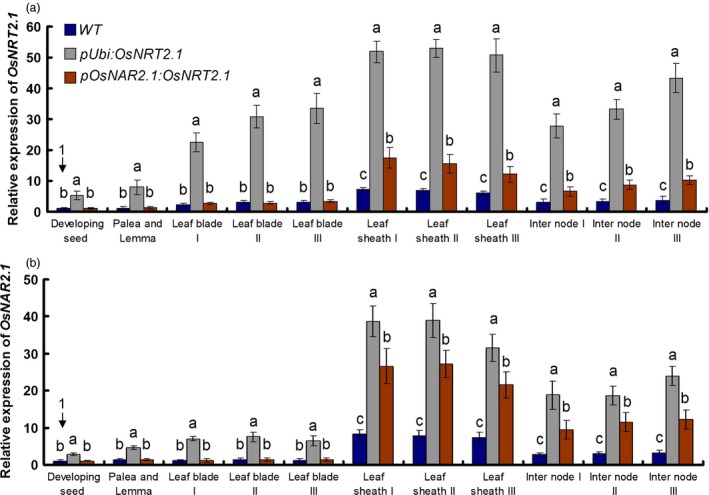
Expression pattern of OsNRT2.1 and OsNAR2.1. Relative expression of (a) OsNRT2.1 and (b) OsNAR2.1 in various organs at 14 days after pollination. pUbi:OsNRT2.1 represents the average of OE1, OE2 and OE3. pOsNAR2.1:OsNRT2.1 represents the average of O6, O7 and O8. Statistical analysis was performed on data derived from the T4 generation. We defined that developing seed of the wild‐type (WT) expression was set equal to 1. Error bars: SE (n = 3). Significant differences between the transgenic lines and WT are indicated by different letters (P < 0.05, one‐way ANOVA).
Compared with WT, the OsNRT2.1 expression increased by about 7.5‐fold averagely in all organs of OE lines including root. The increase pattern of OsNRT2.1 in OE lines showed the similar trade as the native expression of OsNRT2.1 in the WT that was most in root, secondly in leaf sheaths, thirdly in leaf blades and internodes and lest in grain (Table S5, Figure 3a). It was very interesting that we found that in OE lines, the OsNAR2.1 was also increased with the pattern as most in root, secondly in leaf sheaths, thirdly in internodes, fourthly in leaf blades and lest in grain (Table S5, Figure 3b). The coexpression increase pattern of OsNRT2.1 and OsNAR2.1 occurred in all organs of OE lines (Table S5, Figure S7).
Compared with WT, the OsNRT2.1 expression was not changed in grain and leaf blades in O lines and increased in leaf sheaths, internodes and root significantly with a same pattern as WT, which is most in root, secondly in leaf sheaths, thirdly in internodes, fourthly in leaf blades and lest in grain (Table S5, Figure 3a). For OsNAR2.1 expression in O lines, it was also not increased in grain and leaf blades only increased in leaf sheaths, internodes and root significantly with a same pattern as WT, which was most in root, secondly in leaf sheaths, thirdly in internodes and lest in grain and leaf blades (Table S5, Figure 3b). The coexpression increase pattern of OsNRT2.1 and OsNAR2.1 occurred in leaf sheaths, internodes and root of O lines (Table S5, Figure S7).
Expression patterns of OsNRT2.1 and OsNAR2.1 in different growth stages of the WT and transgenic lines
In this study, we found that the OsNRT2.1 and OsNAR2.1 mRNA levels in the culms including the leaf sheath and internode (Figure S8) were significantly higher in all of the transgenic plants than in the WT plants (Figure 4a,b). OsNRT2.1 expression was 3–20‐fold higher in the OE lines than in the WT, but was only 31%–45% higher in the O lines than in the WT (Figure 4a). OsNAR2.1 expression was two‐ to ninefold higher in the OE lines than in the WT and was one‐ to eightfold higher in the O lines than in the WT (Figure 4b). Throughout the experimental growth period, OsNRT2.1 expression was significantly higher in the culms of the OE lines than the O lines, but no significant difference in OsNAR2.1 expression was observed between the OE and O transgenic lines.
Figure 4.
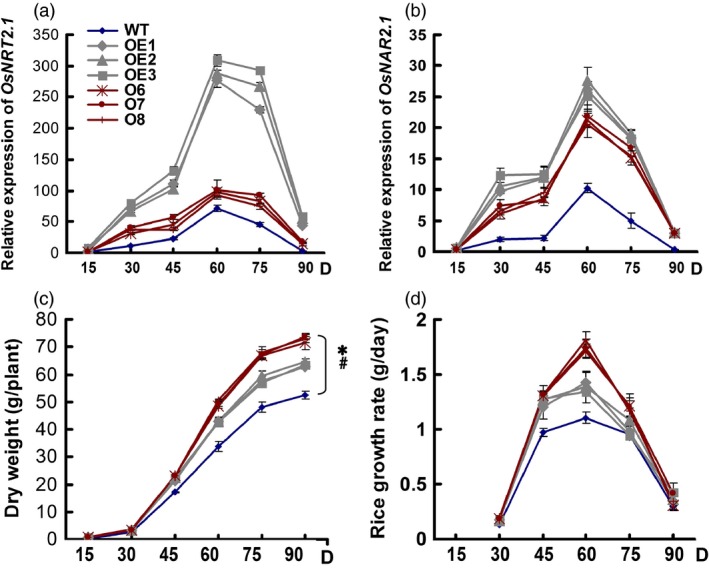
Growth status of the wild‐type (WT) and transgenic lines during the experimental growth period. (a) Changes in OsNRT2.1 expression over the experimental growth period. (b) Changes in OsNAR2.1 expression over the experimental growth period. RNA was extracted from culms. (c) Dry weight. Dry weight mean values are for all aboveground biomass, including the grain yield. (d) Growth rate. Samples were collected at 15‐day intervals after seedlings were transplanted to the field. Statistical analysis was performed on data derived from the T3 generation. Error bars: SE (n = 3). D in x‐axis means the day after transplanting. The asterisk at the end of time course indicates their statistically significant differences among plants, and #statistically significant differences during the growth stages (P < 0.05, ANCOVA).
During the entire experimental growth period, no significant differences in the OsNRT2.1 and OsNAR2.1 expression were found between the leaf blade I of the O lines and WT plants, but the expression levels of both OsNRT2.1 and OsNAR2.1 were up‐regulated significantly in the OE plants relative to the WT (Figure S9).
Growth rate in transgenic lines
N transport and the growth of rice biomass are closely related, and OsNRT2.1 overexpression was previously shown to affect the rice growth (Katayama et al., 2009). In this study, the OE and O lines began to show significantly higher biomass than WT plants at 45 days after transplanting and had accumulated 21% and 38% more biomass at 90 days (Figure 4c). The growth rates of the OE and O lines reached peak values at 60 days and were higher than those of the WT plants (Figure 4d). The growth rates of the OE and O lines were approximately 25% and 58% greater, respectively, than the WT. The growth rates of the transgenic and WT plants were identical after 75 days during the grain filling stage (Figure 4d).
The coexpression of OsNRT2.1 and OsNAR2.1 in the WT and transgenic plants
The expression pattern of OsNRT2.1 and OsNAR2.1 in different organs showed that there exists a strong coexpression pattern of these two genes in rice plants (Figure S7). The coexpression pattern of OsNRT2.1 and OsNAR2.1 was altered very much in OE lines compared with O and WT lines (Figure S7). The expression ratio of OsNRT2.1 and OsNAR2.1 is 5.4 : 1 in the OE organs and 3.6 : 1 in the O lines compared with 3.9 : 1 in the WT organs (Figure S7). Furthermore, we specially investigated the ratio of OsNAR2.1 to OsNRT2.1 expression in root as 6.3 : 1 in the OE lines, 4.1 : 1 in the O lines and 4.2 : 1 in the WT plants, with no significant differences between the O lines and WT plants (Table S5).
The culm is important for N storage and translocation in rice shoots. In rice shoot, OsNRT2.1 and OsNAR2.1 expression was expressed most in leaf sheaths of culm (Figure 3). Our expression data also confirmed that OsNRT2.1 and OsNAR2.1 expression in the culm could play a key role in remobilization. To further study the possible relationship between OsNRT2.1 and OsNAR2.1 expression and rice growth, we compared the ratio of OsNRT2.1 and OsNAR2.1 expression in rice plants. The expression ratio was approximately 11.3 : 1 in the OE lines and approximately 4.7 : 1 in the O lines compared with approximately 7.2 : 1 in the WT plants (Figure 5). We also investigated the ratio of OsNAR2.1 to OsNRT2.1 expression in leaf blade I. The expression ratio was 7.3 : 1 in the OE lines, 4 : 1 in the O lines and 5.2 : 1 in the WT plants, with no significant differences between the O lines and WT plants (Figure S10). The ratio of OsNAR2.1 to OsNRT2.1 expression correlated with the grain yield.
Figure 5.
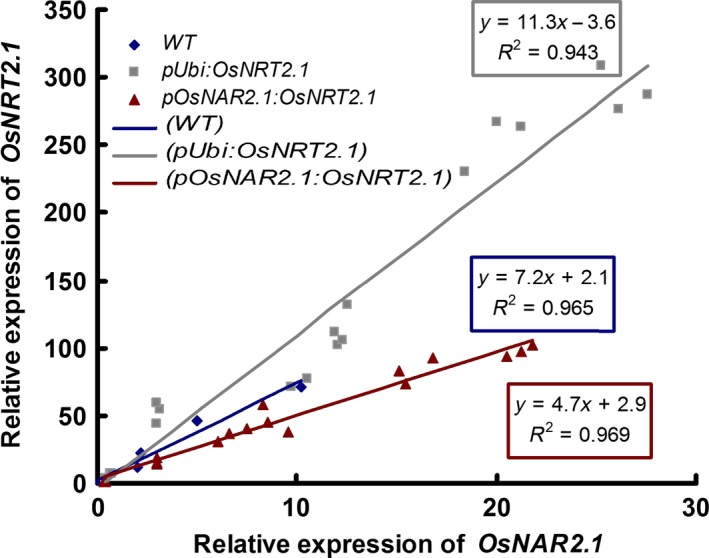
Ratios of OsNRT2.1 to OsNAR2.1 expression in culms of the wild‐type (WT) and transgenic lines over the course of the study. The ratios of OsNRT2.1 and OsNAR2.1 expression during different periods in the culms of pUbi:OsNRT2.1 lines (OE1, OE2 and OE3), pOsNAR2.1:OsNRT2.1 lines (O6, O7 and O8) and WT were presented.
Discussion
N nutrition affects all levels of plant function, from metabolism to resource allocation, growth and development (Crawford, 1995; Scheible et al., 1997, 2004; Stitt, 1999). As one form of available N nutrient to plants, is taken up in the roots by active transport processes and stored in vacuoles in rice shoots (Fan et al., 2007; Li et al., 2008). In rice, OsNAR2.1 acts as a partner protein with OsNRT2.1 in the uptake and transport of (Liu et al., 2014; Tang et al., 2012; Yan et al., 2011). OsNAR2.1 gene expression was shown to be up‐regulated by and down‐regulated by (Feng et al., 2011; Nazoa et al., 2003; Zhuo et al., 1999).
Rooke et al. (2000) reported that the maize Ubi‐1 promoter had strong activity in young, metabolically active tissues and in pollen grains. Furthermore, Cornejo et al. (1993) performed histochemical localization of Ubi‐GUS activity and showed that the Ubi promoter was most active in rapidly dividing cells; however, Chen et al. (2012) reported that the Ubi promoter drove strong OsPIN2 expression in all tissues. Chen et al. (2015) reported that ectopic expression of the WOX11 gene driven by the promoter of the OsHAK16 gene, which encodes a potassium (K) transporter that is induced by low K levels, led to an extensive root system, adventitious roots and increased tiller numbers in rice. In contrast, WOX11 overexpression driven by the Ubi promoter induced ectopic crown roots in rice and failed to present any similar super growth phenotype in field (Zhao et al., 2009) as described by Chen et al. (2015). These results suggested that the use of a specific inducible promoter‐driven gene function could be a good strategy for plant breeding.
In this study, OsNRT2.1 expression was up‐regulated significantly in both the aboveground and underground parts of pUbi:OsNRT2.1 transgenic plants relative to the WT (Figure 1c), while OsNRT2.1 expression in pOsNAR2.1:OsNRT2.1 transgenic plants was increased significantly only in the roots and culms and not enhanced significantly in the leaves (Figure 1d). Specific induction of expression by the OsNAR2.1 promoter in rice roots and culms based on GUS fusion data has been reported previously (Feng et al., 2011); therefore, we investigated the effects of tissue‐specific induction of OsNRT2.1 expression in roots and culms on plant growth and nitrogen‐use efficiency (NUE).
Effect of pOsNAR2.1:OsNRT2.1 expression on NUE in transgenic rice
N redistribution during the reproductive stage was shown to vary significantly among cultivars and under various N management strategies (Souza et al., 1998). Mae and Ohira (1981) reported that a major proportion of N was redistributed from vegetative organs to panicles during grain filling, 64% of which was derived from leaf blades and 36% from culms. The NTE values of the WT, pUbi:OsNRT2.1 and pOsNAR2.1:OsNRT2.1 plants were averagely 49.5%, 33.4% and 50.3%, indicating that N transfer from the shoots into grain was significantly less in pUbi:OsNRT2.1 transgenic plants than in the WT or pOsNAR2.1:OsNRT2.1 plants (Table 4). This lower level of N transfer from vegetative organs to grain during grain filling in pUbi:OsNRT2.1 plants affected spike formation and final grain yield compared with the WT and pOsNAR2.1:OsNRT2.1 plants (Table 1). The DMTE values for WT, pUbi:OsNRT2.1 and pOsNAR2.1:OsNRT2.1 plants were 22.1%, 5.5% and 22.1%, averagely (Table 4) demonstrating that markedly less dry matter was transferred into grain yield in the pUbi:OsNRT2.1 lines. These data confirmed that the transport of N and biomass during the transition from the flowering to harvest stages affected the final yield and NUE of rice (Zhang et al., 2009) and also indicated that the Ubi promoter decreased N and biomass translocation, while the OsNAR2.1 promoter did not.
In both types of OsNRT2.1 overexpression line, NT was reduced during the reproductive stage and NUE was reduced before flowering. The CPAY average values of the WT, pUbi:OsNRT2.1 and pOsNAR2.1:OsNRT2.1 plants were 28.5%, 11% and 34.9%, respectively. The CPAY of the pOsNAR2.1:OsNRT2.1 plants was higher than that of the WT plants that had higher CPAY than the pUbi:OsNRT2.1 plants (Table 4). The HI was much lower for the pUbi:OsNRT2.1 plants than for the WT or pOsNAR2.1:OsNRT2.1 plants (Table 4), indicating that the Ubi promoter affected uptake and N use before the flowering stage and that levels of OsNRT2.1 overexpression in rice that were excessive did not benefit N use during either the vegetative or reproductive stages.
The coexpression pattern of OsNRT2.1 and OsNAR2.1 is an important factor controlling N transport in rice
How to assess the effect of transporter expression on rice NUE is a key question for rice breeding. The transporter, OsNRT1.1B, was shown to improve the NUE of rice by approximately 30% (Hu et al., 2015), while our data showed that not the higher expression level of transporter was relative to the higher yield and NUE of rice (Tables 1 and 4, and Figure 4). After determining the expression levels of OsNRT2.1 and its partner gene, OsNAR2.1, we calculated the coexpression ratio of these genes in rice plants.
The coexpression pattern of OsNRT2.1 and OsNAR2.1 happened in the WT and transgenic plants (Figures 3 and 4, Table S5). However, the coexpression pattern of OsNRT2.1 and OsNAR2.1 was changed in OE lines compared with O and WT lines (Figure S7), which suggested that OsNRT2.1 driven by different promoters had a different coexpression patterns with OsNAR2.1. But it is still not clear that why increasing OsNRT2.1 expression would induce OsNAR2.1 expression and what mechanism exists behind the coexpression pattern of OsNRT2.1 and OsNAR2.1 in the gene regulation.
However, the ratio changes of OsNRT2.1 to OsNAR2.1 expression may be a clue for the explanation of the rice growth and nitrogen‐use difference in the WT and transgenic lines. The ratio changes of OsNRT2.1 to OsNAR2.1 expression in different organs were increased significantly in pUbi:OsNRT2.1 lines compared with WT and pOsNAR2.1:OsNRT2.1 lines (Figure S7). Also during the growth stages, the ratio of OsNRT2.1 to OsNAR2.1 expression in culm (including the internode and leaf sheath) was increased in pUbi:OsNRT2.1 lines compared with WT and the pOsNAR2.1:OsNRT2.1 lines (Figure 5). These data indicated that the interaction between OsNRT2.1 and OsNAR2.1 in pUbi:OsNRT2.1 plants differed from WT and that in the pOsNAR2.1:OsNRT2.1 lines. Furthermore in culms, pOsNAR2.1:OsNRT2.1 lines showed a lower expression ratio of these two genes, in which more OsNAR2.1 protein may be available to interact with OsNRT2.1 protein. Therefore, the efficiency of OsNRT2.1 function in rice plants should differ between the two types of transgenic plants resulting in different rice yield and NUE phenotypes. On the other hand, the higher expression of OsNRT2.1 and OsNAR2.1 in all the organs of pUbi:OsNRT2.1 than WT may cause some disadvantages to plants such as high cost for mRNA synthesis or disturbing of nitrogen transport in the leaf blades. All possibilities remain to be confirmed by further analysis.
In this study, we showed that the rice yield and NUE could be improved by increasing OsNRT2.1 expression, especially in combination with a lower expression ratio with its partner gene OsNAR2.1, which encodes a high‐affinity transporter.
Experimental procedures
Construction of vectors and rice transformation
We amplified the OsNRT2.1/OsNRT2.2 ORF sequence, which is identical for both genes, from cDNA isolated from Oryza sativa L. ssp. Japonica cv. Nipponbare using the primers listed in Table S1. We amplified the OsNAR2.1 and ubiquitin promoters from the pOsNAR2.1(1698bp):GUS (Feng et al., 2011) and pUbi:OsPIN2 (Chen et al., 2012) constructs, respectively, using the primers listed in Table S2. The PCR products were cloned into the pMD19‐T vector (TaKaRa Biotechnology, Dalian, China) and confirmed by restriction enzyme digestion and DNA sequencing. The pUbi:OsNRT2.1 and pOsNAR2.1:OsNRT2.1 vectors were constructed as shown in Figure S1. These constructs were introduced into Agrobacterium tumefaciens strain EHA105 by electroporation and then transformed into rice as described previously (Tang et al., 2012).
Southern blot analysis
Transgene copy number was determined by the Southern blot analysis following procedures described previously (Jia et al., 2011). Briefly, genomic DNA was extracted from the leaves of wild type (WT) and digested with HindIII and EcoRI restriction enzymes. The digested DNA was separated on a 1% (w/v) agarose gel, transferred to a Hybond‐N+ nylon membrane and hybridized with hygromycin‐resistant gene.
Biomass, total nitrogen (N) measurement and calculation of NUE
WT and transgenic rice plants were harvested at 9:00 a.m. and heated at 105 °C for 30 min. Panicles, leaves and culms were then dried at 75 °C for 3 days. Dry weights were recorded as biomass values. Samples collected at 15‐day intervals from WT and transgenic lines grown in soil in pots were used to calculate whole‐plant biomass values.
Total N content was measured using the Kjeldahl method (Li et al., 2006). The total dry weight (biomass) was estimated as the sum of weights of all plant parts. Total N accumulation was estimated as the sum of the N contents of all plant parts. Agronomic NUE (ANUE, g/g) was calculated as (grain yield − grain yield of zero N plot)/N supply; NRE (%) was calculated as (total N accumulation at maturity for N‐treated plot − total N accumulation at maturity of zero N plot)/N supply; physiological NUE (PNUE, g/g) was calculated as (grain yield − grain yield of zero N plot)/total N accumulation at maturity; and the NHI (%) was calculated as (grain N accumulation at maturity/total N accumulation at maturity). Dry matter and NT and translocation efficiency method for the calculation of the reference in Ntanos and Koutroubas (2002) and Zhang et al. (2009). Dry matter translocation (DMT, g/m2) was calculated as dry matter at anthesis − (dry matter at maturity − grain yield); dry matter translocation efficiency (DMTE, %) was calculated as (dry matter translocation/dry matter at anthesis) × 100%; the CPAY (%) was calculated as (dry matter translocation/grain yield) × 100%; the HI (%) was calculated as (grain yield/dry matter at maturity) × 100%; PANU (g/m2) was calculated as total N accumulation at maturity − total N accumulation at anthesis; NT (g/m2) was calculated as total N accumulation at anthesis − (total N accumulation at maturity − grain N accumulation at maturity); NTE (%) was calculated as (N translocation/total N accumulation at anthesis) × 100%; and the CPNGN (%) was calculated as (N translocation/grain N accumulation at maturity) × 100% (Table S4).
The growth conditions for T0 to T4 transgenic plants
T0, T2, T3 and T4 generation plants were grown in plots at the Nanjing Agricultural University in Nanjing, Jiangsu (Figure S11). T1 generation plants were grown in Sanya, Hainan. Jiangsu is in a subtropical monsoon climate zone. Chemical properties of the soils in the plots at the Nanjing Agricultural University included organic matter, 11.56 g/kg; total N content, 0.91 g/kg; available P content, 18.91 mg/kg; exchangeable K, 185.67 mg/kg; and pH 6.5. Basal applications of 30 kg P/ha (Ca(H2PO4)2) and 60 kg/K ha (KCl) were made to all plots 3 days before transplanting. N fertilizer accounted for 40%, 30% and 40% of the total N fertilizer was applied prior to transplanting, at tillering, just before the heading stage, respectively.
The field experiments for yield harvest
T0–T4 generation seedlings were planted in the same experiment site in Nanjing, except T1 in Sanya. Seed generation transgenic lines and WT were surface‐sterilized with 10% (v : v) hydrogen peroxide (H2O2) for 30 min and rinsed thoroughly with deionized water. The transgenic seeds were soaked in water containing 25 mg/L hygromycin, and the WT seeds were soaked in water. After 3 days, the sterilized seeds were sown evenly in wet soil. The similar seedlings were transplanted to field plots after 3 weeks of germination.
T1–T3 plants were planted in plots fertilized at a rate of 300 kg N/ha as urea and in plots without N fertilization. Plots were 2 × 2.5 m in size with the seedlings planted in a 10 × 10 array. Plants at the edges of all four sides of each plot were removed at maturity to avoid the influence of edge effects. Four points, each containing four seedlings, totally 16 seedlings, were selected randomly within the remaining centre 8 × 8 array of plants, and samples were collected (Khuram et al., 2013; Ookawa et al., 2010; Pan et al., 2013; Srikanth et al., 2016). Yield and biomass values determined from these four points in each plot were used to calculate the yield per hectare and biomass of each line, and three random plots for each line were designed in the experiment (Figure S11).
T3 generation plants were sampled at 15‐day intervals for the determination of the grain yield, biomass and N content. The growth rate was the dry weight of the weight increase in the unit time after seedlings were transplanted to the plots.
T4 generation plants were planted in a plot fertilized at a rate of 0, 180 and 300 kg N/ha as urea. Same random field plots with three replicates were designed as T1–T3 plants for yield, and biomass values determined from these four points were used to calculate the yield and biomass per plant and ANUE of each line.
mRNA sampling and qRT‐PCR assay
To investigate the expression pattern in plant organs, we sampled mRNA for seeds, palea and lemma, leaf blade I, leaf blade II, leaf blade III, leaf sheath I, leaf sheath II, leaf sheath III, internode I, internode II, internode III and newly developed root (3 cm from root tips) at the grain filling stage (described in Figure S6). Tracking rice in the whole growth period of gene expression in T3 generation, we sampled mRNA from culms including leaf sheath and internode I (described in Figure S8) at 15, 30, 45, 60, 75 and 90 days after transplanting.
Total RNAs were prepared from the various tissues of the WT and transgenic plants using TRIzol reagent (Vazyme Biotech Co., Ltd, http://www.vazyme.com). Real‐time PCR was carried as described before (Li et al., 2014). All primers used for qRT‐PCR are listed in Table S3.
Statistical analysis
Data were analysed by Tukey's test of one‐way analysis of variance (ANOVA), except that analysis of covariate (ANCOVA) was used in the biomass and growth rate during growth stages (Figure 4a,b). Different letters on the histograms or after mean values indicate statistically significant differences at P < 0.05 between the transgenic plants and WT (one‐way ANOVA). The asterisk at the end of time course indicates their statistically significant differences among plants, and #statistically significant differences during the growth stages at P < 0.05 (ANCOVA). All statistical evaluations were conducted using the IBM SPSS Statistics version 20 software (SPSS Inc., Chicago, IL).
Supporting information
Figure S1 Diagram of (a) pUbi:OsNRT2.1 and (b) pOsNAR2.1:OsNRT2.1 constructs.
Figure S2 Characterization of T0 generation transgenic lines.
Figure S3 Grain yield and dry weight of WT and T1 generation transgenic plants.
Figure S4 Southern blot analysis of transgene copy number.
Figure S5 Grain yield, dry weight and ANUE of WT and T4 generation transgenic plants under low and normal N supplies.
Figure S6 The diagram of RNA sampling in T4 generation transgenic lines and WT plants.
Figure S7 Ratios of OsNRT2.1 to OsNAR2.1 expression in different organs of WT and transgenic lines.
Figure S8 The diagram of RNA sampling in T3 generation transgenic lines and WT plants.
Figure S9 Changes in genes expression in leaf blade I throughout the experimental growth period.
Figure S10 Ratios of OsNRT2.1 and OsNAR2.1 expression in the leaf blade I of WT and transgenic plants during different periods.
Figure S11 A field experiment picture of WT and T3 generation transgenic plants.
Table S1 Primers used to amplify the OsNRT2.1 open reading frame.
Table S2 Primers used to amplify the OsNAR2.1 and Ubiquitin promoters.
Table S3 Primers used to detect OsActin, OsNAR2.1, and OsNRT2.1 gene expression.
Table S4 Methods of NUE calculations.
Table S5 Real‐time quantitative RT‐PCR analysis of endogenous OsNRT2.1 and OsNAR2.1 expression in various transgenic lines and wild‐type (WT) plants.
Acknowledgements
This work was supported by the National Natural Science Foundation (No. 31372122, 31172013) and Nanjing 321 Talents Program. The English in this document has been checked by two native speakers of English, and for a certificate, please see: http://www.textcheck.com/certificate/SoEIdm.
References
- Chen, Y. , Fan, X. , Song, W. , Zhang, Y. and Xu, G. (2012) Over‐expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 10, 139–149. [DOI] [PubMed] [Google Scholar]
- Chen, G. , Feng, H. , Hu, Q. , Qu, H. , Chen, A. , Yu, L. and Xu, G. (2015) Improving rice tolerance to potassium deficiency by enhancing OsHAK16p:WOX11‐controlled root development. Plant Biotechnol. J. 13, 833–848. [DOI] [PubMed] [Google Scholar]
- Cornejo, M.J. , Luth, D. , Blankenship, K.M. , Anderson, O.D. and Blechl, A.E. (1993) Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 23, 567–581. [DOI] [PubMed] [Google Scholar]
- Coruzzi, G. and Bush, D.R. (2001) Nitrogen and carbon nutrient and metabolite signaling in plants. Plant Physiol. 125, 61–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coruzzi, G.M. and Zhou, L. (2001) Carbon and nitrogen sensing and signaling in plants: emerging ‘matrix effects’. Curr. Opin. Plant Biol. 4, 247–253. [DOI] [PubMed] [Google Scholar]
- Crawford, N.M. (1995) Nitrate: nutrient and signal for plant growth. Plant Cell, 7, 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M. and Forde, B.G. (2002) Molecular and developmental biology of inorganic nitrogen nutrition In The Arabidopsis Book (Meyerowitz E.M., ed.), pp. 1–25. Rockville, MD: American Society of Plant Biologists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M. and Glass, A.D.M. (1998) Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 3, 389–395. [Google Scholar]
- Fan, X. , Shen, Q. , Ma, Z. , Zhu, H. , Yin, X. and Miller, A.J. (2005) A comparison of nitrate transport in four different rice (Oryza sativa L.) cultivars. Sci. China C Life Sci. 48, 897–911. [PubMed] [Google Scholar]
- Fan, X. , Jia, L. , Li, Y. , Smith, S.J. , Miller, A.J. and Shen, Q. (2007) Comparing nitrate storage and remobilization in two rice cultivars that differ in their nitrogen use efficiency. J. Exp. Bot. 58, 1729–1740. [DOI] [PubMed] [Google Scholar]
- Feng, Q. , Zhang, Y. , Hao, P. , Wang, S. , Fu, G. , Huang, Y. , Li, Y. et al (2002) Sequence and analysis of rice chromosome 4. Nature, 420, 316–320. [DOI] [PubMed] [Google Scholar]
- Feng, H. , Yan, M. , Fan, X. , Li, B. , Shen, Q. , Miller, A.J. and Xu, G. (2011) Spatial expression and regulation of rice high‐affinity nitrate transporters by nitrogen and carbon status. J. Exp. Bot. 62, 2319–2332. [DOI] [PubMed] [Google Scholar]
- Hu, B. , Wang, W. , Ou, S. , Tang, J. , Li, H. , Che, R. , Zhang, Z. et al (2015) Variation in NRT1.1B contributes to nitrate‐use divergence between rice subspecies. Nat. Genet. 47, 834–838. [DOI] [PubMed] [Google Scholar]
- Jia, H. , Ren, H. , Gu, M. , Zhao, J. , Sun, S. , Zhang, X. , Chen, J. et al (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156, 1164–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama, H. , Mori, M. , Kawamura, Y. , Tanaka, T. , Mori, M. and Hasegawa, H. (2009) Production and characterization of transgenic rice plants carrying a high‐affinity nitrate transporter gene (OsNRT2.1). Breed. Sci. 59, 237–243. [Google Scholar]
- Khuram, M. , Asif, I. , Muhammad, H. , Faisal, Z. , Siddiqui, M.H. , Mohsin, A.U. , Bakht, H.F.S.G. et al (2013) Impact of nitrogen and phosphorus on the growth, yield and quality of maize (Zea mays L.) fodder in Pakistan. Philipp. J. Crop Sci. 38, 43–46. [Google Scholar]
- Kirk, G.J.D. and Kronzucker, H.J. (2005) The potential for nitrification and nitrate uptake in the rhizosphere of wetland plants: a modelling study. Ann. Bot. 96, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker, H.J. , Glass, A.D.M. , Siddiqi, M.Y. and Kirk, G.J.D. (2000) Comparative kinetic analysis of ammonium and nitrate acquisition by tropical lowland rice: implications for rice cultivation and yield potential. New Phytol. 145, 471–476. [DOI] [PubMed] [Google Scholar]
- Li, B. , Xin, W. , Sun, S. , Shen, Q. and Xu, G. (2006) Physiological and molecular responses of nitrogen‐starved rice plants to re‐supply of different nitrogen sources. Plant Soil, 287, 145–159. [Google Scholar]
- Li, Y.L. , Fan, X.R. and Shen, Q.R. (2008) The relationship between rhizosphere nitrification and nitrogen‐use efficiency in rice plants. Plant Cell Environ. 31, 73–85. [DOI] [PubMed] [Google Scholar]
- Li, Y. , Gu, M. , Zhang, X. , Zhang, J. , Fan, H. , Li, P. , Li, Z. et al (2014) Engineering a sensitive visual tracking reporter system for real‐time monitoring phosphorus deficiency in tobacco. Plant Biotechnol. J. 12, 674–684. [DOI] [PubMed] [Google Scholar]
- Liu, X. , Huang, D. , Tao, J. , Miller, A.J. , Fan, X. and Xu, G. (2014) Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two‐component system for high‐affinity nitrate transport. New Phytol. 204, 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mae, T. and Ohira, K. (1981) The remobilization of nitrogen related to leaf growth and senescence in rice plants (Oryza sativa L.). Plant Cell Physiol. 22, 1067–1074. [Google Scholar]
- Nazoa, P. , Vidmar, J.J. , Tranbarger, T.J. , Mouline, K. , Damiani, I. , Tillard, P. , Zhuo, D. et al (2003) Regulation of the nitrate transporter gene AtNRT2.1 in Arabidopsis thaliana: responses to nitrate, amino acids, and developmental stage. Plant Mol. Biol. 52, 689–703. [DOI] [PubMed] [Google Scholar]
- Ntanos, D.A. and Koutroubas, S.D. (2002) Dry matter and N accumulation and translocation for Indica and Japonica rice under Mediterranean conditions. Field. Crop. Res. 74, 93–101. [Google Scholar]
- Okamoto, M. , Kumar, A. , Li, W. , Wang, Y. , Siddiqi, M.Y. , Crawford, N.M. and Glass, A.D. (2006) High‐affinity nitrate transport in roots of Arabidopsis depends on expression of the NAR2‐like gene AtNRT3.1. Plant Physiol. 140, 1036–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ookawa, T. , Hobo, T. , Yano, M. , Murata, K. , Ando, T. , Miura, H. , Asano, K. et al (2010) New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 1, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsel, M. , Chopin, F. , Leleu, O. , Smith, S.J. , Krapp, A. , Daniel‐Vedele, F. and Miller, A.J. (2006) Characterization of a two‐component high‐affinity nitrate uptake system in Arabidopsis. Physiology and protein–protein interaction. Plant Physiol. 142, 1304–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, S. , Rasul, F. , Li, W. , Tian, H. , Mo, Z. , Duan, M. and Tang, X. (2013) Roles of plant growth regulators on yield, grain qualities and antioxidant enzyme activities in super hybrid rice (Oryza sativa L.). Rice, 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, A. , Galvan, A. and Fernandez, E. (1994) Identification of nitrate transporter genes in Chlamydomonas reinhardtii . Plant J. 5, 407–419. [DOI] [PubMed] [Google Scholar]
- Rooke, L. , Byrne, D. and Salgueiro, S. (2000) Marker gene expression driven by the maize ubiquitin promoter in transgenic wheat. Ann. Appl. Biol. 136, 167–172. [Google Scholar]
- Sasakawa, H. and Yamamoto, Y. (1978) Comparison of the uptake of nitrate and ammonium by rice seedlings. Plant Physiol. 62, 665–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, T. , Matsumoto, T. , Yamamoto, K. et al (2002) The genome sequence and structure of rice chromosome 1. Nature, 420, 312–316. [DOI] [PubMed] [Google Scholar]
- Scheible, W.R. , Gonzalez‐Fontes, A. , Lauerer, M. , Muller‐Rober, B. , Caboche, M. and Stitt, M. (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell, 9, 783–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible, W.R. , Morcuende, R. , Czechowski, T. , Fritz, C. , Osuna, D. , Palacios‐Rojas, N. , Schindelasch, D. et al (2004) Genome‐wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 136, 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza, S.R. , Stark, E.M.L.M. and Fernandes, M.S. (1998) Nitrogen remobilization during the reproductive period in two Brazilian rice varieties. J. Plant Nutr. 21, 2049–2063. [Google Scholar]
- Srikanth, B. , Subhakara, R.I. , Surekha, K. , Subrahmanyam, D. , Voleti, S.R. and Neeraja, C.N. (2016) Enhanced expression of OsSPL14 gene and its association with yield components in rice (Oryza sativa) under low nitrogen conditions. Gene, 576, 441–450. [DOI] [PubMed] [Google Scholar]
- Stitt, M. (1999) Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 2, 178–186. [DOI] [PubMed] [Google Scholar]
- Tang, Z. , Fan, X. , Li, Q. , Feng, H. , Miller, A.J. , Shen, Q. and Xu, G. (2012) Knockdown of a rice stelar nitrate transporter alters long‐distance translocation but not root influx. Plant Physiol. 160, 2052–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Guegler, K. , LaBrie, S.T. and Crawford, N.M. (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell, 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Fan, X. and Miller, A.J. (2012) Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Yan, M. , Fan, X. , Feng, H. , Miller, A.J. , Sheng, Q. and Xu, G. (2011) Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant, Cell Environ. 34, 1360–1372. [DOI] [PubMed] [Google Scholar]
- Yong, Z. , Kotur, Z. and Glass, A.D. (2010) Characterization of an intact two‐component high‐affinity nitrate transporter from Arabidopsis roots. Plant J. 63, 739–748. [DOI] [PubMed] [Google Scholar]
- Zhang, H. and Forde, B.G. (2000) Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 51, 51–59. [PubMed] [Google Scholar]
- Zhang, Y.L. , Fan, J.B. , Wang, D.S. and Shen, Q.R. (2009) Genotypic differences in grain yield and physiological nitrogen use efficiency among rice cultivars. Pedosphere, 19, 681–691. [Google Scholar]
- Zhao, Y. , Hu, Y. , Dai, M. , Huang, L. and Zhou, D.X. (2009) The WUSCHEL‐related homeobox gene WOX11 is required to activate shoot‐borne crown root development in rice. Plant Cell, 21, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo, D. , Okamoto, M. , Vidmar, J.J. and Glass, A.D. (1999) Regulation of a putative high‐affinity nitrate transporter (Nrt2;1At) in roots of Arabidopsis thaliana . Plant J. 17, 563–568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Diagram of (a) pUbi:OsNRT2.1 and (b) pOsNAR2.1:OsNRT2.1 constructs.
Figure S2 Characterization of T0 generation transgenic lines.
Figure S3 Grain yield and dry weight of WT and T1 generation transgenic plants.
Figure S4 Southern blot analysis of transgene copy number.
Figure S5 Grain yield, dry weight and ANUE of WT and T4 generation transgenic plants under low and normal N supplies.
Figure S6 The diagram of RNA sampling in T4 generation transgenic lines and WT plants.
Figure S7 Ratios of OsNRT2.1 to OsNAR2.1 expression in different organs of WT and transgenic lines.
Figure S8 The diagram of RNA sampling in T3 generation transgenic lines and WT plants.
Figure S9 Changes in genes expression in leaf blade I throughout the experimental growth period.
Figure S10 Ratios of OsNRT2.1 and OsNAR2.1 expression in the leaf blade I of WT and transgenic plants during different periods.
Figure S11 A field experiment picture of WT and T3 generation transgenic plants.
Table S1 Primers used to amplify the OsNRT2.1 open reading frame.
Table S2 Primers used to amplify the OsNAR2.1 and Ubiquitin promoters.
Table S3 Primers used to detect OsActin, OsNAR2.1, and OsNRT2.1 gene expression.
Table S4 Methods of NUE calculations.
Table S5 Real‐time quantitative RT‐PCR analysis of endogenous OsNRT2.1 and OsNAR2.1 expression in various transgenic lines and wild‐type (WT) plants.


