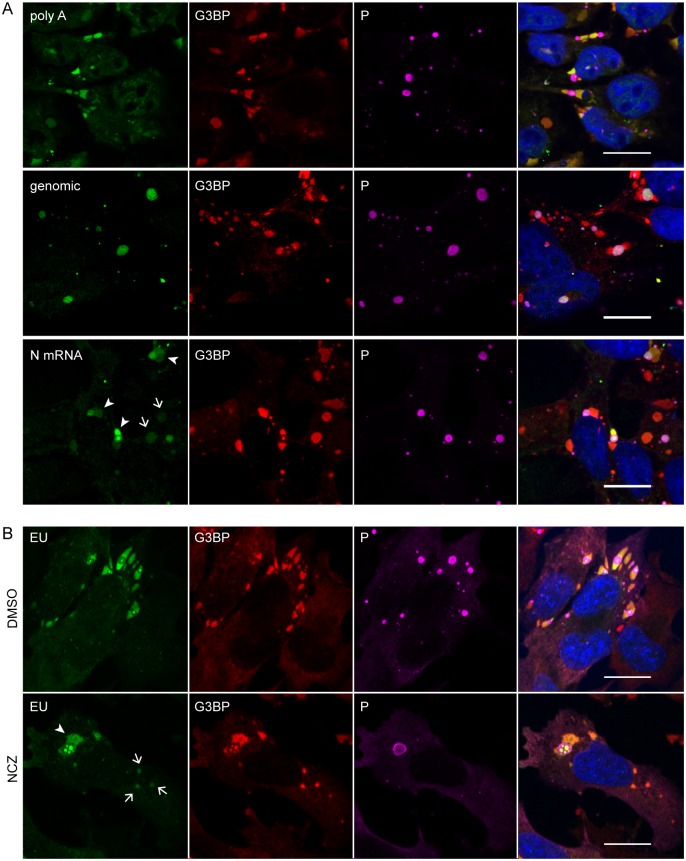
Fig 8. Viral mRNAs are specifically recruited to SGs and their transport from NBs to SGs does not require microtubules.
(A) Cells were infected (MOI = 3) for 24 h. FISH experiments were performed by using 5’-ATTO448 modified oligonucleotides (Eurofins Genomics) to detect cellular mRNA (polyA), viral genomic RNA and N mRNA as described in Material and Methods. The proteins G3BP1 (red) and P (purple) were stained with the mouse anti-G3BP and rabbit anti-P antibodies followed by incubation with secondary fluorescent IgG. DAPI (blue) was used to stain the nuclei (merge). Note that viral mRNAs are detected in SGs close to NBs (arrowheads), but also in SGs far from NBs (arrows). The scale bars correspond to 15 μm. (B) U373-MG were infected for 16 h in presence of nocodazole (2 μM; NCZ) or mock-treated during viral infection. At 16 h p.i, cells were treated with 20 μM Act D and 1mM 5-ethynyl uridine (EU; Invitrogen) for 3 h. Cells were co-stained with the rabbit anti-P (purple) and mouse anti-G3BP (red) antibodies followed by incubation with secondary fluorescent IgG. Viral RNA was visualized by EU labeling of cells (green) as described in Materials and Methods. DAPI (blue) was used to stain the nuclei (merge). Note that viral mRNA are detected in SGs close NBs (arrowheads) but also in SGs far from NBs (arrows). The scale bars correspond to 15 μm.