Fig 3.
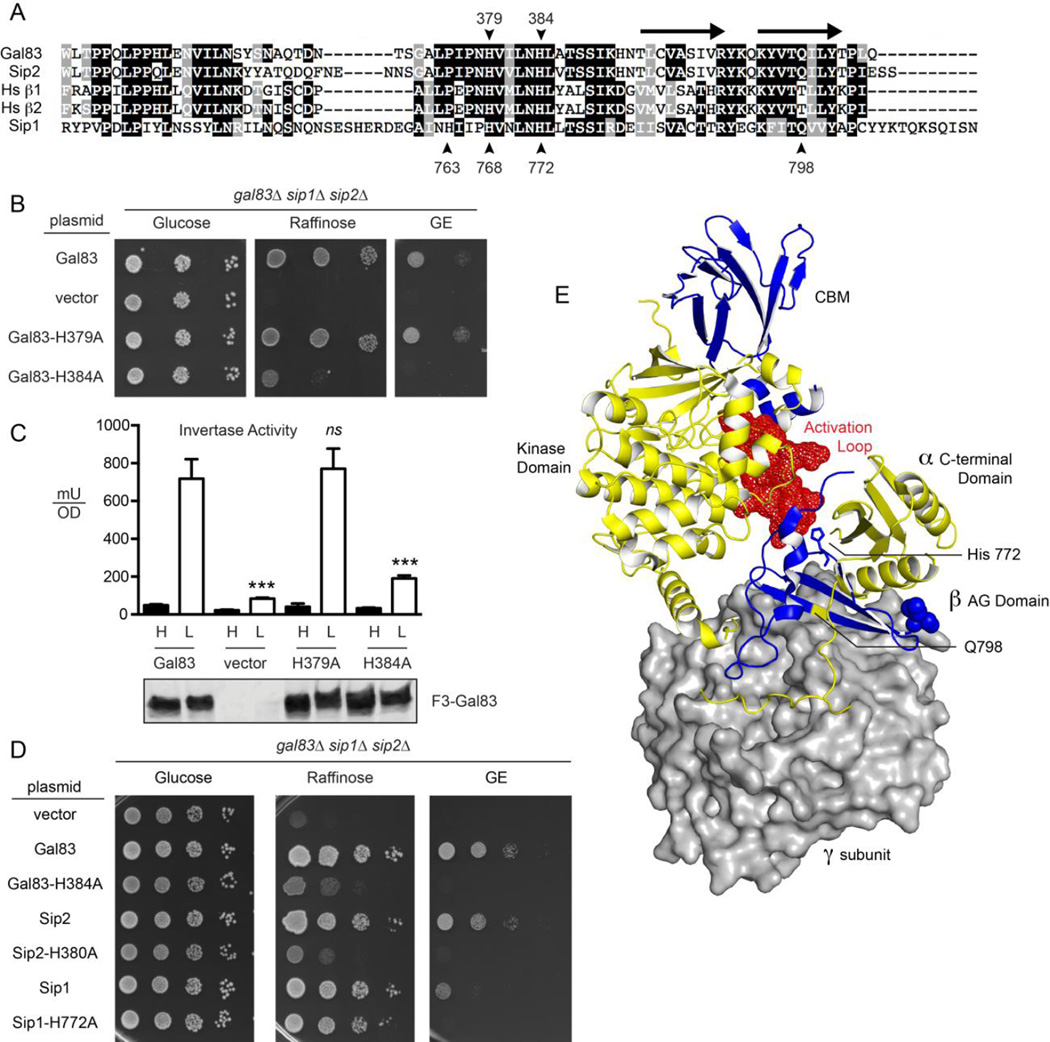
Role of conserved histidine residues in yeast β subunits. (A) Sequence alignment of the α-γ interaction domains of the three β subunits from S. cerevisiae and two β subunits from human (Hs β1 and Hs β2). Positions of the conserved histidine residues are indicated with the numbering on top referring to amino acids in Gal83 and the bottom referring to the Sip1 protein. The position of two β sheets that form the interface between the γ subunit and the C-terminal domain of the α subunit are indicated with arrows above the alignment. The position of Q798 in Sip1 is also indicated. (B) Spot dilution assay to measure β subunit function in yeast lacking all three β subunits but transformed with a low copy plasmid expressing no β subunit (vector), Gal83 or Gal83 with histidine to alanine substitutions at positions 379 or 384. Growth was measured on plates with the glucose, raffinose or a mixture of glycerol and ethanol (GE) as the carbon sources. (C) Invertase enzyme activity was measured in triplicate samples with the mean plotted ±SE. Values statistically different from wild type are indicated. The cells assayed were the same as those used in panel B. Western blot of Flag-tagged Gal83 proteins is shown below. (D) Spot dilution assay to measure β subunit function in yeast lacking all three β subunits but transformed with a low copy plasmid expressing no β subunit (vector), or the β subunit indicated with or without the indicated histidine to alanine substitutions. Growth was measured on plates with the glucose, raffinose or a mixture of glycerol and ethanol (GE) as the carbon sources. (E) Structural model of the Snf1 kinase heterotrimer based on the mammalian AMPK heterotrimer [33]. The γ subunit is shown in surface representation (gray). The α and β subunits are show in the cartoon representation in yellow and blue, respectively. The α subunit kinase domain, C-terminal domain and activation loop (red spheres) are indicated. The β subunit C-terminus and carbohydrate-binding motif (CBM) are indicated. The position of the Sip1 histidine residue 772 is shown in the stick representation while residue glutamine 798 is indicated in yellow. This structural model was created with PyMol (Schrödinger) using the Protein Database file 4CFF.