Abstract
The family of inverse BAR (I-BAR) domain proteins participates in a range of cellular processes associated with membrane dynamics and consists of five distinct members. Three of the I-BAR proteins, including insulin receptor tyrosine kinase substrate (IRTKS), contain an SH3 domain near their C-termini. Yet, the function of the SH3 domain of IRTKS remains uncharacterized. Here we report that in contrast to MIM, which is a prototype of I-BAR proteins and does not contain an SH3 domain, IRTKS promoted serum-induced cell migration along with enhanced phosphorylation of mitogen activated kinases Erk1/2 and p38, and activation of small GTPases Rac1 and Cdc42. In addition, cells overexpressing IRTKS exhibited an increased polarity characterized by elongated cytoplasm and extensive lamellipodia at leading edges. However, a mutant with deletion of the SH3 domain attenuated both cellular motility and p38 phosphorylation but had little effect on Erk1/2 phosphorylation. Also, a chimeric mutant in which the N-terminal portion of MIM is fused with the C-terminal IRTKS, including the SH3 domain, was able to promote chemotactic response to serum and cellular polarity. In contrast, a chimeric mutant in which the N-terminal IRTKS is fused with the C-terminal MIM failed to do so. Furthermore, treatment of cells with SB203580, a selective inhibitor of p38, also neutralized the effect of IRTKS on cell migration. These data indicate that the SH3 domain distinguishes the function of IRTKS in promoting cell migration and inducing signal transduction from those of SH3-less I-BAR proteins.
Keywords: IRTKS, MIM, SH3, p38, small GTPases, cell migration, cell morphology
1. Introduction
Cell migration is fundamental to development, wound healing, cell-cell communication and immune surveillance, and is associated with extensive membrane dynamics, including extension at leading edges, generation of new adhesion sites, and detachment of adhesions at cell rear position [1]. Many of these cellular processes are subjected to the control by Rho-like small GTPases, such as Rac1 and Cdc42, which direct the process of cell polarization and directional migration through their effects on the assembly of the actin cytoskeleton [2]. Also, MAP kinases Erk1/2 and p38 have been implicated in the migration of various types of cells [3]. In addition to these well-characterized signaling molecules, There is emerging evidence for the important role of a protein family that shares a Bin-Amphiphysin-Rvs (BAR) domain in cell migration and cell shape changes [4]. The BAR domain is a dimeric motif that binds to phospholipid membranes through a curved interface. Depending on the shape of the interface, the BAR proteins can be divided into three subfamilies, classical BAR, F-BAR and I-BAR domain proteins. While the former two subfamilies induce membrane invagination, the I-BAR domain has a concave shape and induces membrane protrusion [5]. The mammalian genomes contain five I-BAR domain genes, encoding IRSp53, MIM, ABBA, IRTKS and PINKBAR, respectively [6]. Among them, IRTKS, IRSp53 and PINKBAR are distinguished from others by having an SH3 domain. The best characterized function of the I-BAR protein-associated SH3 domain is the one with IRSp53, which interacts with proline rich sequences of a wide range of actin cytoskeleton-associated proteins, including WAVE2 [7], Mena [8], Dia1 [9] and Eps8 [10]. As these proteins are implicated in the assembly of actin filaments in the cortex such as lamellipodia at cell leading edges, the SH3 domain provides presumably a functional link between IRSp53 and the actin dynamics.
In contrast to IRsp53, the function of the SH3 domain of IRTKS, which is also known as brain-specific angiogenesis inhibitor 1-associated protein 2-like 1 (BAIAP2L1), remains elusive. Although IRTKS is closely related to IRSp53, its SH3 domain is not apparently required for its recruitment to lamellipodia [11]. Nevertheless, IRTKS may use its SH3 domain to link to the actin cytoskeleton as evidenced by its ability to Escherichia coli secreted F-like protein encoded on prophage U, which triggers the actin assembly to facilitate bacterial invasion into host cells [12]. In addition, IRTKS is implicated in plasma membrane dynamics and actin bundling associated with cell shape changes, migration and proliferation [5]. Mice with depletion of the IRTKS gene showed insulin resistance symptoms such as glucose intolerance, hyperglycemia, insulin less-sensitivity, hyperinsulinemia, and excessive production of hepatic glucose [13]. Furthermore, IRTKS suppresses innate immune responses against RNA virus through the Rig-IMAVs signaling pathway, thereby down-regulating extravagant inflammation [14]. IRTKS is also involved in tumor progression and has been reported to inhibit the p53 induced apoptosis by direct regulating its transcriptional activity [15]. However, there is little known about the cellular function of IRTKS that is relevant to these reported phenomena.
As an effort to understand the distinct function of I-BAR proteins, we compared IRTKS with MIM and found that while MIM inhibits cell migration in response to serum, IRTKS promotes chemotactic response and cell polarization. Importantly, we found that the IRTKS-mediated cell migration and polarization is dependent upon its SH3 domain. Thus, the SH3 domain is the primary motif that functionally distinguishes I-BAR domain proteins.
2. Materials and methods
2.1 Cell culture and DNA transfection
HeLa cells were cultured under 5% CO2, 37°C in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, NY) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and 100 unit/ml Penicillin and Streptomycin. DNA transfection was performed with cells at 90% confluence by using FuGene HD Transfection Reagent (Active Motif, Carlsbad, CA) with the protocol recommended by manufacturer’s instruction. Stable transfected cells were selected in the medium containing 0.5 mg/ml G418 for two weeks with constant medium changes every three days. The selected cells were pooled together and used for all the analyses as described.
2.2 Plasmids
Plasmids encoding MIM-GFP and GFP were prepared as described previously [16]. pIRTKS-GFP was synthesized and cloned into the vector pEZ-M98 by Genecopoeia (Gaithersburg, MD). pIRTKSΔSH3, which encodes a protein with a deletion of the SH3 domain (aa 342-399), was prepared by using a QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) using pIRTKS-GFP as the template. To prepare pIRTKS-I-BAR-MIM-CT-GFP, a DNA sequence that has an open reading frame corresponding to the human IRTKS protein from aa 1 to 231 and human MIM protein from aa 235 to 755 was synthesized by GeneScript (Nanjing, China) and cloned into pUC57 vector. This DNA sequence is also flanked by a HindIII site at the 5′ and a BamH1 site at the 3′, respectively. After digestion with HindIII and BamH1, the insert was subcloned into pEZ-M98. To prepare pMIM-I-BAR-IRTKS-CT-GFP, a DNA sequence that is flanked by HindIII and BamH1 sites and has an open reading frame corresponding to human MIM protein from aa 1 to 250 and human IRTKS protein from aa 250 to 511 was synthesized by GeneScript. This DNA fragment was also subcloned into pEZ-M98 as above. For all the mutants, the fidelity of the mutations was confirmed by DNA sequencing.
2.3 Analysis of phosphorylated p38 and Erk1/2
The level of phosphorylated MAP Kinases p38 and Erk1/2 was measured by Western blot assay using antibodies against specifically pp38 and pErk1/2, as described previously [3].
2.4 Rac1 and Cdc42 activation assay
To measure GTP-Rac1 and GTP-Cdc42, equal amount of cell lysates was precipitated with 20 μl 50% GST-PAK-CRIB agarose beads (Millipore, MA). Samples were separated by 15% SDS-PAGE and subjected to Western blot using antibodies against Rac1 or Cdc42 [3].
2.5 Cell migration assay
Cell migration was evaluated by Transwell assay as described previously [3].
2.6 Analysis of cell morphology
Images of cells were captured by digital camera QImaging MicroPublisher 5.0 RTV that is equipped on Nikon Eclipse TE2000-U microscope and controlled by MetaMorph software. Cell elongation factor was determined according to Frances [17]. Briefly, the long axis was defined as the longest length of the cell, and the length across the nucleus in a direction perpendicular to the long axis was recognized as the short axis. The elongation factor was defined as the ratio of two axes. Both axes were measured by ImageJ software.
2.7 Statistical analysis
All the data were subjected to two tailed Student’s t-test by using GraphPad Prism software. A difference with a P value < 0.01 was considered as statistically significant.
3. Results
3.1 IRTKS promoted cell migration in response to serum
To evaluate the effect of IRTKS on cell motility, cells expressing IRTKS-GFP and cells expressing GFP only were subjected to Transwell assay for chemotactic response to serum at different concentrations for various times. In the absence of serum, less than 2% of both cell types was migrated in 16 hr (Fig. 1A). In the presence of serum, both control cells and IRTKS expressing cells showed significant motility responses in a dose dependent manner, which reached to a maximal level at 15% serum (Fig 1A). However, the number of migrated IRTKS-GFP cells was nearly 2 and half of that of control cells at the maximal level. When cell migration towards 15% serum was analyzed for different times, IRTKS-GFP cells also showed a greater migration rate than did control cells during the period from 8 to 24 hr (Fig. 1B). As a comparison, we also analyzed cells expressing GFP-tagged MIM protein. As shown in Fig 1C and 1D, MIM-GFP cells showed a lower response to serum under the same condition compared with cells expressing GFP only. This result is consistent with our previous finding that overexpression of MIM often inhibits cell migration [18].
Fig. 1. IRTKS promotes cell motility.
(A, B) HeLa cells expressing IRTKS-GFP or GFP were migrated through Transwell plates towards FBS at various concentrations for 16 hr (A) or towards 15% FBS for various times (B). (C, D) HeLa cells expressing MIM-GFP or GFP cells were also analyzed by Transwell assay for chemotactic response to FBS at varying concentrations (C) or to 15% FBS for different times (D). (E) Schematic presentation of IRTKS, MIM, and IRTKS mutants. CT, C-terminal; NT, N-terminal; I-BAR, inverse BAR domain; PRD, proline rich domain; and W, WH2 domain. (F) Comparison of the motility response of cells expressing MIM and different IRTKS mutants to 15% FBS. ***, P<0.001, n=3.
The differential response to serum with IRTKS and MIM overexpressors prompted us to seek the structural basis for their functional difference. Compared to MIM, IRTKS is distinguished by having an SH3 domain at the C-terminal (CT) domain (Fig. 1E). To examine whether the CT domain is responsible for the difference, we switched the N-terminal domain (NT) and the CT domain of MIM and IRTKS each other, resulting in two chimeric mutants: MIM-I-BAR-IRTKS-CT-GFP, and IRTKS-I-BAR-MIM-CT-GFP. HeLa cells expressing each mutant were analyzed by Transwell assay for motility response to 15% FBS for different times. As shown in Fig. 1F, cells expressing MIM-I-BAR-IRTKS-CT-GFP displayed a response at a level comparable to that of IRTKS-GFP cells. In contrast, IRTKS-I-BAR-MIM-CT-GFP cells reduced motility response as did MIM-GFP cells. To further confirm the role of the SH3 domain in cell migration, we also generated a mutant IRTKSΔSH3-GFP, which had a deletion of the SH3 domain. When the cells expressing the mutant were analyzed for the response to serum by Transwell, we found that the cells also showed a poor mobility response (Fig 1F). Hence, the SH3 domain determines the response of cells expressing I-BAR proteins to serum.
3.2 IRTKS promoted activation of MAPKs, Rac1 and Cdc42
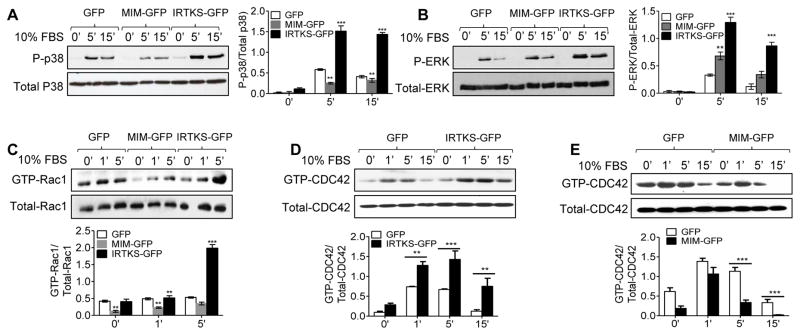
We next examined several signaling molecules commonly involved in cell migration, including MAP kinases p38 and Erk1/2, and small GTPases Rac1 and Cdc42. In response to serum stimulation, phosphorylation of p38 in cells expressing IRTKS-GFP was greatly enhanced by nearly 3 folds within 15 min compared with that in cells expressing GFP only (Fig. 2A). In contrast, the level of p38 phosphorylation in the cells expressing MIM-GFP after exposure to serum was about half of that in control cells (Fig. 2A). However, both IRTKS-GFP and MIM-GFP expressing cells displayed remarkable increase in phosphorylation of Erk1/2 although the increase was more dramatic in IRTKS-GFP cells (Fig. 2B). Likewise, the serum-induced increase in GTP-Rac1 and GTP-Cdc42 was more significant in IRTKS-GFP cells than in control cells (Fig. 2C and 2D). On the other hand, MIM-GFP cells showed less increase in the levels of GTP-Rac1 and GTP-Cdc42 than those in control cells under the same condition (Fig. 2C and 2E). Thus, IRTKS promoted the activation of these signaling molecules, whereas MIM suppressed them except of Erk1/2.
Fig. 2. IRTKS regulates MAPKs and small GTPases upon serum treatment.
HeLa Cells expressing IRTKS-GFP, MIM-GFP or GFP were stimulated by 10% FBS for the indicated times, and then subjected to examination for p38 phosphorylation (A), Erk1/2 phosphorylation (B) and Rac1 activation. Cdc42 activation was also examined in cells expressing IRTKS-GFP or GFP (D), MIM-GFP or GFP (E) after treated with 10% FBS for the indicated times. Quantification of these activated proteins was based on normalization to the total amount of individual these proteins. **, P<0.0.1, and ***, P<0.001, n=3.
3.3 The SH3 domain is responsible for p38 phosphorylation and cell migration
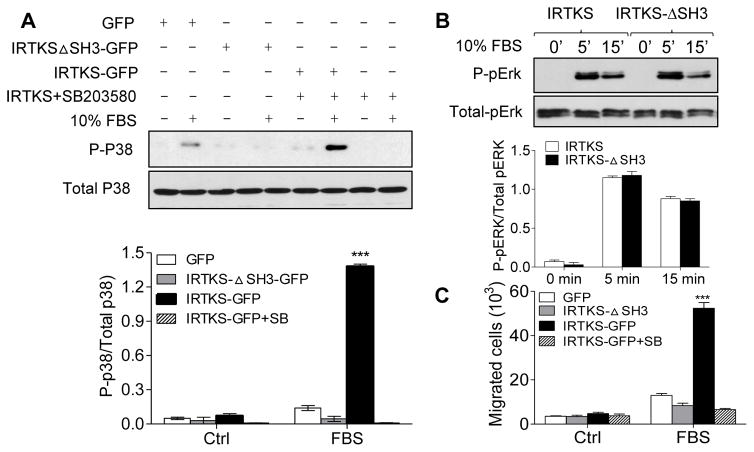
We were interested in the role of the SH3 domain in the IRTKS-mediated signal transduction with focusing on p38 and Erk1/2 by examining cells expressing IRTKSΔSH3-GFP cells. As shown in Fig 3A, overexpression of IRTKSΔSH3-GFP nearly abolished the phosphorylation of p38 in response to serum. In contrast, the mutant showed no effect on the phosphorylation of Erk1/2 (Fig. 3B), suggesting that the SH3 domain is more involved in activation of p38 than Erk1/2. To examine any functional relationship between p38 and IRTKS-mediated cell migration, we treated cells expressing IRTKS-GFP with SB203580, a compound that inhibits both p38a and p38b, prior to serum stimulation. This treatment blocked not only p38 phosphorylation (Fig 3A) but also inhibited serum-mediated cell migration to the degree similar to that as shown with the cells expressing IRTKSΔSH3-GFP (Fig 3C). Therefore, the SH3 domain promoted IRTKS-mediated cell migration in a p38 phosphorylation dependent manner.
Fig. 3. The SH3 domain is indispensable for IRTKS-mediated p38 phosphorylation.
(A) Hela cells expressing GFP, IRTKS-GFP, and IRTKSΔSH3-GFP were treated with 10% FBS for 5 min and subjected to Western blot analysis for the level of phosphorylated p38. As a negative control, IRTKS-GFP cells were pre-treated with 10 μM SB203580 1 hr prior to adding serum. (B) Phosphorylation of Erk1/2 in cells expressing IRTKS-GFP or IRTKSΔSH3-GFP was evaluated after treated with 10% FBS for the indicated times. (C) Cells expressing IRTKS mutants were analyzed for the chemotactic response to serum in the presence and absence of 10 μM SB203580. ***, P<0.001, n=3.
3.4 IRTKS induced cell shape changes in a manner depending upon its SH3 domain
I-BAR proteins are known to induce profoundly cell shape changes, and thus we inspected HeLa cells expressing MIM-GFP and different IRTKS mutants after staining with phalloidin, a chemical that stains specifically stress fibers. Compared to control cells expressing GFP only (Fig. 4A), cells expressing either IRTKS-GFP or MIM-GFP displayed significantly shape changes. Over 90% of MIM-GFP cells exhibited extensive filopodia-like protrusions (white arrow heads), whereas over 90% of cells expressing IRTKS-GFP displayed elongated cytoplasm with extensive lamellipodia at the leading edge (Fig 4A, yellow arrow head). However, cells expressing IRTKSΔSH3-GFP failed to show the similar elongated shape (Fig 4B). Also, we analyzed cells expressing MIM-I-BAR-IRTKS-CT-GFP, which apparently exhibited elongated cytoplasm as well as filopodia-like extension but few lamellipodia at leading edges (Fig 4B). In contrast, cells expressing IRTKS-I-BAR-MIM-CT-GFP showed neither extensive lamellipodia at cell leading edges nor elongated cytoplasm (Fig. 4B). Quantification of shape changes based on elongation factor demonstrated that cells expressing IRTKS-GFP had an elongation factor more than twice as that of control cells, whereas cells expressing MIM-GFP had a smallest elongation factor (Fig. 4C). Also, cells expressing IRTKS-I-BAR-MIM-CT-GFP and IRTKSΔSH3-GFP had a small elongation factor, whereas cells expressing MIM-I-BAR-IRTKS-CT-GFP had an elongation factor that was about 1.5 folds of that of control cells (Fig 4C). Taken together, the SH3 domain plays an important role in the IRTKS-mediated cell shape changes.
Fig. 4. The SH3 domain is required for IRTKS-mediated cell shape changes.
HeLa cells expressing GFP, MIM-GFP, and IRTKS-GFP (A) or HeLa cells expressing MIM-I-BAR-IRTKS-CT-GFP, IRTKS-I-BAR-MIM-CT-GFP and IRTKSΔSH3-GFP (B) were cultured in serum-containing medium and co-stained with GFP antibody and Alexa flour 594 tagged phalloidin. The stained cells were inspected by fluorescent microscopy. Bars: 80 μm. (C) Quantification of the degree of cell elongation as described in the Materials and Methods. The data shown is mean ± SEM of three independent experiments. In each experiment, 50 cells were analyzed. ***, P<0.001.
4. Discussion
In this presented study we showed that overexpression of IRTKS had a dramatic impact on serum-induced chemotaxis, activations of p38, Erk1/2, Rac1 and Cdc42, and cell shape changes. The finding that IRTKS promotes cell migration is consistent with other reports showing that IRTKS was overexpressed in certain metastatic lesions and that IRTKS upregulated the motility of HT1080 cancer cells [19]. Significantly, we found that the SH3 domain is indispensable for IRTKS-mediated cell migration. To our knowledge, this is the first report about a functional link between the SH3 domain of IRTKS and cell migration. Although the direct partners for the SH3 domain of IRTKS have not yet been characterized, the SH3 domain of IRSp53, which is closely related to IRTKS, is known to bind to Wiskott-Aldrich syndrome protein (WASP) and WASP-family verprolin-homologous (WAVE) proteins [20]. Both WASP and WAVE proteins are the activators for the Arp2/3 complex, the primary actin nucleator responsible for the assembly of the branched actin filaments that are enriched in the lamellipodia at cell leading edges, which are often associated with the initiation of cell migration. We speculate that the SH3 domain of IRTKS may have a similar function to interact with an actin cytoskeleton associated protein that is involved in actin reorganization, and thereby promoting cell migration. In contrast, MIM does not have an SH3 domain. Instead, it has a proline-rich domain that is known to bind to cortactin, which also has an activity to activate the Arp2/3 complex by stabilizing its association with actin filaments [21]. However, the function of MIM in the actin assembly varies significantly depending on the Arp2/3 complex activators. While MIM promotes cortactin/Arp2/3-mediated actin assembly, it inhibits N-WASP/Arp2/3-mediated actin polymerization [18]. Since N-WASP is the primary Arp2/3 complex activator, overexpression of MIM often results in inhibition of cell migration. Recently, we also reported that MIM is implicated in the internalization of chemokine receptor CXCR4 [3], the pathway that leads to downregulation of CXCR4 signaling. While it is unclear about the primary factor(s) in the serum responsible for the observed chemotaxis of HeLa cells, MIM could also promote its endocytosis as well, thereby inhibiting the motility response to them.
Deletion of the SH3 domain also led to the failure to induce p38 phosphorylation, which is necessary for the serum-mediated cell migration. Interestingly, the SH3 domain was not required for enhanced Erk1/2 phosphorylation in response to serum, indicating that different mechanisms are used by IRTKS to enhance these two MAPKs and that p38 phosphorylation, rather than Erk1/2 phosphorylation, is responsible for the IRTKS-mediated cell migration. Yet, the nature for these mechanisms is currently unknown although it is speculated that a common feature shared by IRTKS and MIM may be responsible for Erk1/2 phosphorylation. Indeed, the I-BAR domains of both IRTKS and MIM are known to bind to small GTPase Rac1 [22,23], activation of which could lead to activation of Erk1/2. In addition to Rac1, IRTKS was recently reported to interact with small GTPase Rif by its I-BAR domain [24]. Hence, it is quite possible that IRTKS and MIM could bind to different small GTPases, which may influence profoundly their subsequent signaling under different cellular contexts.
In addition to cell migration, we found that overexpression of IRTKS and MIM induced different cell shape changes. While overexpression of MIM promotes the formation of filopodia-like microspikes, overexpression of IRTKS caused cellular polarization with extremely extended the cytoplasm and increased lamellipodia at the leading edges. Again, the formation of such polarity is dependent upon the SH3 domain. Of note, the chimeric mutant with N-terminal MIM and C-terminal IRTKS also showed increased cell polarity but without extensive lammellipodia formation, suggesting that there is a coordination between the SH3 domain and the I-BAR domain, and that different I-BAR domains have different functions. Indeed, IRTKS binds membranes mainly based on electrostatic interaction, whereas an amphipathic helix in MIM I-BAR is inserted into membrane bilayers, facilitating more efficient membrane bending [25]. These differences could explain specific function of each I-BAR protein. For example, IRTKS induces dorsal membrane ruffles [24] and is associated with invadopodia in cancer cells and podosomes in osteoclasts [26]. In contrast, MIM induces filopodia-like protrusions when it is overexpressed [27]. These differences highlight a necessity to characterize the physiological function of each I-BAR protein under different cellular contexts.
Supplementary Material
Highlights.
Expression of IRTKS enhanced cell migration towards serum;
IRTKS modulated phosphorylation of p38 and pERK1/2 MAPKs as well as Rho GTPase pathways in response to stimulation of serum;
Src homology 3 (SH3) domain in IRTKS is responsible for its effects on p38 phosphorylation, which is indispensable for cell motility.
SH3 domain of IRTKS is also responsible for its effect on elongated cell shape;
Acknowledgments
We are grateful to Dr. Tailan Zhan and Dr. Yanbao Xiong for their helps on experiments. This research is supported by grant number R01CA113809 from the National Cancer Institute of the NIH (to X.Z.), China National High-tech Research and Development Project (863 Project, 2013AA032205, to M.J.), Foundation of China for Key Project of International Cooperation (61420106012 to N. G), and the Maryland Stem Cell Research Fund (2012-0081 to X.Z.).
Footnotes
6. Author Contribution
L.L. and X. Z. designed the project, analyzed the data, interpreted the results and wrote the manuscript with inputs from other authors. H.L supplied some materials used for mutagenesis, involved in carrying out some of experiments. S.B. analyzed, interpreted the results and made suggestions to the manuscript preparation. N.G. and M.J. were the mentors of L.L. in China and provided general input on the project.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Zhan T, Cao C, Li L, Gu N, Civin C, Zhan X. MIM regulates the trafficking of bone marrow cells via modulating surface expression of CXCR4. Leukemia. 2016 doi: 10.1038/leu.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell reports. 2013;4:1213–1223. doi: 10.1016/j.celrep.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang YP, Huang LY, Sun WM, Zhang ZZ, Fang JZ, Wei BF, Wu BH, Han ZG. Insulin receptor tyrosine kinase substrate activates EGFR/ERK signalling pathway and promotes cell proliferation of hepatocellular carcinoma. Cancer Lett. 2013;337:96–106. doi: 10.1016/j.canlet.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Zhao H, Pykäläinen A, Lappalainen P. I-BAR domain proteins: linking actin and plasma membrane dynamics. Curr Opin Cell Biol. 2011;23:14–21. doi: 10.1016/j.ceb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T. Optimization of WAVE2 complex–induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. The Journal of cell biology. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krugmann S, Jordens I, Gevaert K, Driessens M, Vandekerckhove J, Hall A. Cdc42 induces filopodia by promoting the formation of an IRSp53: Mena complex. Curr Biol. 2001;11:1645–1655. doi: 10.1016/s0960-9822(01)00506-1. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara T, Mammoto A, Kim Y, Takai Y. Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem Biophys Res Commun. 2000;271:626–629. doi: 10.1006/bbrc.2000.2671. [DOI] [PubMed] [Google Scholar]
- 10.Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8–IRSp53 complex. Nat Cell Biol. 2006;8:1337–1347. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- 11.Robens JM, Yeow-Fong L, Ng E, Hall C, Manser E. Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14-3-3 binding and SH3-mediated localization. Mol Cell Biol. 2010;30:829–844. doi: 10.1128/MCB.01574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin YS, Yue XL, Zhang QY, Wu XY, Cao Z, Dai ZF. Cerasomal doxorubicin with long-term storage stability and controllable sustained release. Acta Biomater. 2012;8:3372–3380. doi: 10.1016/j.actbio.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 13.Huang LY, Wang YP, Wei BF, Yang J, Wang JQ, Wu BH, Zhang ZZ, Hou YY, Sun WM, Hu RM. Deficiency of IRTKS as an adaptor of insulin receptor leads to insulin resistance. Cell Res. 2013;23:1310–1321. doi: 10.1038/cr.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia P, Wang S, Xiong Z, Ye B, Huang L-Y, Han Z-G, Fan Z. IRTKS negatively regulates antiviral immunity through PCBP2 sumoylation-mediated MAVS degradation. Nat Commun. 2015;6 doi: 10.1038/ncomms9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang KS, Chen G, Shen HL, Li TT, Chen F, Wang QW, Wang ZQ, Han ZG, Zhang X. Insulin receptor tyrosine kinase substrate enhances low levels of MDM2-mediated p53 ubiquitination. PLoS ONE. 2011;6:e23571. doi: 10.1371/journal.pone.0023571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Zhou K, Zeng X, Lin J, Zhan X. Tyrosine phosphorylation of missing in metastasis protein is implicated in platelet-derived growth factor-mediated cell shape changes. J Biol Chem. 2007;282:7624–7631. doi: 10.1074/jbc.M608448200. [DOI] [PubMed] [Google Scholar]
- 17.McWhorter FY, Wang T, Nguyen P, Chung T, Liu WF. Modulation of macrophage phenotype by cell shape. Proceedings of the National Academy of Sciences. 2013;110:17253–17258. doi: 10.1073/pnas.1308887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin J, Liu J, Wang Y, Zhu J, Zhou K, Smith N, Zhan X. Differential regulation of cortactin and N-WASP-mediated actin polymerization by missing in metastasis (MIM) protein. Oncogene. 2005;24:2059–2066. doi: 10.1038/sj.onc.1208412. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Li T, Zhang L, Yi M, Chen F, Wang Z, Zhang X. Src-stimulated IRTKS phosphorylation enhances cell migration. FEBS Lett. 2011;585:2972–2978. doi: 10.1016/j.febslet.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Takenawa T, Suetsugu S. The WASP–WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- 21.Uruno T, Liu J, Zhang P, Fan Y-x, Egile C, Li R, Mueller SC, Zhan X. Activation of Arp2/3 complex-mediated actin polymerization by cortactin. Nat Cell Biol. 2001;3:259–266. doi: 10.1038/35060051. [DOI] [PubMed] [Google Scholar]
- 22.Millard TH, Dawson J, Machesky LM. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J Cell Sci. 2007;120:1663–1672. doi: 10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- 23.Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118:5393–5403. doi: 10.1242/jcs.02640. [DOI] [PubMed] [Google Scholar]
- 24.Sudhaharan T, Sem KP, Liew HF, Yu YH, Goh WI, Chou AM, Ahmed S. Rho GTPase Rif signals through IRTKS, Eps8 and WAVE2 to generate dorsal membrane ruffles and filopodia. J Cell Sci. 2016 doi: 10.1242/jcs.179655. jcs. 179655. [DOI] [PubMed] [Google Scholar]
- 25.Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19:95–107. doi: 10.1016/j.cub.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 26.Oikawa T, Matsuo K. Possible role of IRTKS in Tks5-driven osteoclast fusion. Commun Integr Biol. 2012;5:511–515. doi: 10.4161/cib.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao M, Zhan TL, Ji M, Zhan X. Dimerization is necessary for MIM-mediated membrane deformation and endocytosis. Biochem J. 2012;446:469–475. doi: 10.1042/BJ20120329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.