Figure 7.
MyT1 and Notch Pathways Oppositely Regulate Many Transcriptional Targets
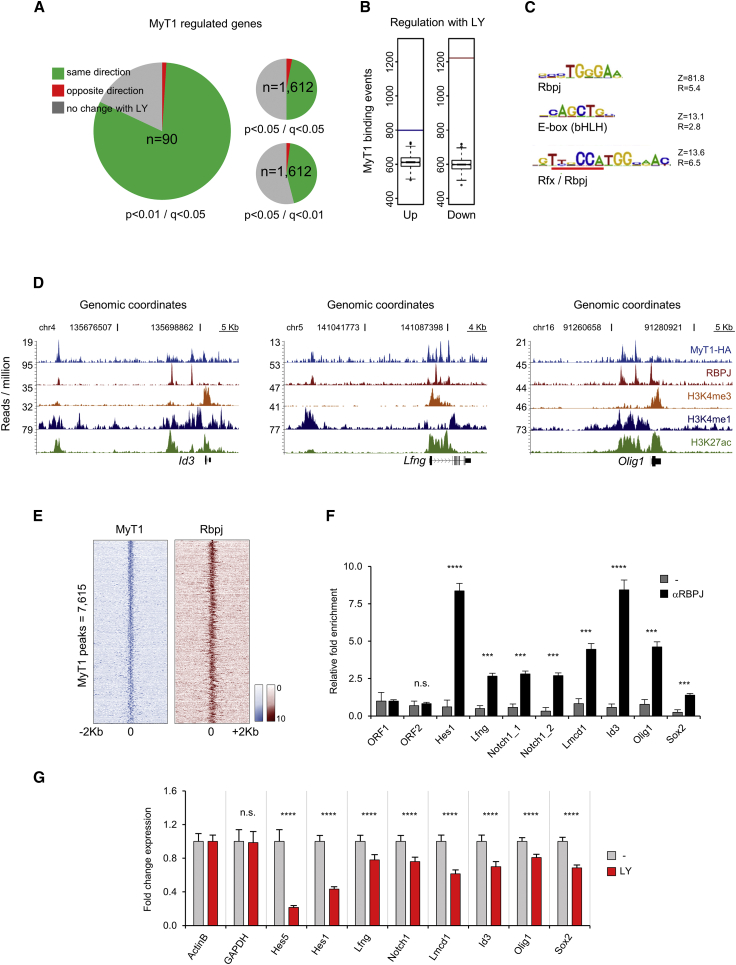
(A) Pie charts representing the percentage of genes deregulated in NS5 cells 4 hr upon MyT1 GoF and deregulated in the same direction (green), opposite direction (red), or unchanged/data unavailable (gray) 4 hr upon LY treatment. Cutoff values for analysis upon MyT1 GoF (p value) and exposure to LY (q value) are indicated in figure next to each pie chart. n, total number of genes.
(B) Number of MyT1 BEs associated with genes that are up- (blue bar) or downregulated (red bar) upon LY treatment. Boxplot distributions of MyT1-binding associations, which can be found testing against 1,000 different random sets of genes, are shown. Test data are represented as box with median of test and first and third quartiles (whiskers, ±1.5× IQR).
(C) Top overrepresented motifs in 50-bp regions centered at MyT1 peak summits are shown. Z, Z score; R, enrichment ratio.
(D) Examples of MyT1 (blue) and RBPJ (red) ChIP-seq enrichment profiles at vicinity of common bound genes and associated H3K4me3 (orange), H3K4me1 (dark blue), and H3K27ac (green) ChIP-seq enrichment profiles are shown.
(E) Density plot of MyT1 (blue) and RBPJ (red) ChIP-seq reads at 4-kb genomic regions centered at MyT1 peak summits (signal intensity represents normalized tag count) is shown.
(F) Validation of RBPJ binding to selected genes by ChIP-qPCR in NS5 cells is shown. ORF1 and ORF2, negative control regions. Mean ± SD of triplicate assays.
(G) Validation of gene expression changes of selected genes in NS5 cells 4 hr after LY treatment by expression qPCR is shown.
Data are shown as mean ± SD; n.s., p > 0.05; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001, according to Student’s t test (F and G). See also Figure S6.