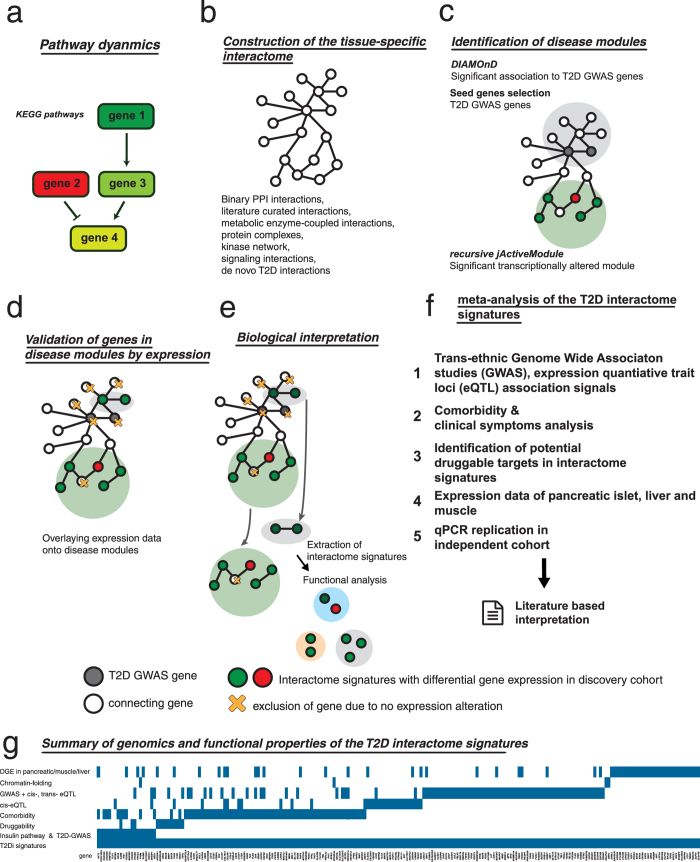
Figure 1. Overview of the integrated T2D study design.
(a) Analysis of pathway dynamics in T2D. (b) T2D interactome was constructed from curation of known interactions and T2D co-expression patterns. (c) Disease modules were identified through identification of genes significantly associated with T2D-GWAS loci, and gene clusters which were significantly altered transcriptionally in the discovery case-control cohort. (d) The genes in the disease modules were filtered and validated based on differential gene expression in our dataset which yielded the final interactome signatures. (e) The resultant interactome signatures were interpreted using a functional network. (f) The interactome signatures were validated through comparative analysis with DIAGRAM GWAS, eQTL studies including MuTHER and GTEx, trait and druggability analysis, expression in pancreatic islet, liver and muscle, followed by qPCR replication of genes in an independent case-control cohort. Refer to respective section for details. (g) T2D interactome signatures overlapping with various genomic and functional properties are defined as follows: “DGE in T2D pancreatic/muscle/liver” indicates the signatures were also dysregulated in T2D relevant tissues, in addition to our discovery cohort; “Chromatin-folding” indicates the genes which may be distantly regulated by the GWAS SNPs reaching genome-wide significance in DIAGRAM-database; “GWAS + cis-, trans-eQTL” refers to the genes identified by the Sherlock statistical framework to be associated with T2D using the DIAGRAM-database, cis- and trans- eQTL signals; “cis-eQTL” refers to the SNPs in perfect LD to T2D interactome signatures with eQTL properties regulating these genes; “Comorbidity” refers to genes that are shared between T2D and comorbid diseases; “Druggability” indicates T2D druggable or potentially druggable targets; “Insulin & T2D-GWAS” refers to genes in the insulin pathway and T2D genes in GWAS Catalogue with dysregulation in the T2D interactome signatures.