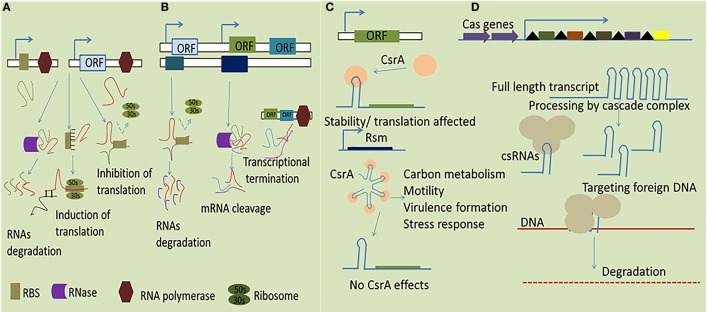
Figure 1.
Simplified representation of mechanisms by which sRNAs function in bacteria. (A) The trans-encoded sRNAs interact with their specific target through imperfect base pairing which ultimately results in both positive and negative effect in altering gene expression to promote RNase degradation of the double-stranded RNA molecules. (B) The cis-encoded sRNAs share extensive complementarity by binding with target mRNA resulting in transcriptional termination, degradation of the sRNA–target RNA complex, and affecting translation through a putative loop formation and, consequently, the cessation of the RNA polymerase activity. (C) The dimeric RNA-binding protein CsrA interacts with target mRNA, typically represented in a hairpin loop structure, transcription termination, leading to an alteration of the accessibility of the translation machinery, and/or the stability of the RNA. (D) Mechanism of action of CRISPR arrays for transcription full length RNA which directly target the foreign DNA via CAS proteins resulting in subsequent degradation of the exogenous DNA.