FIG 9.
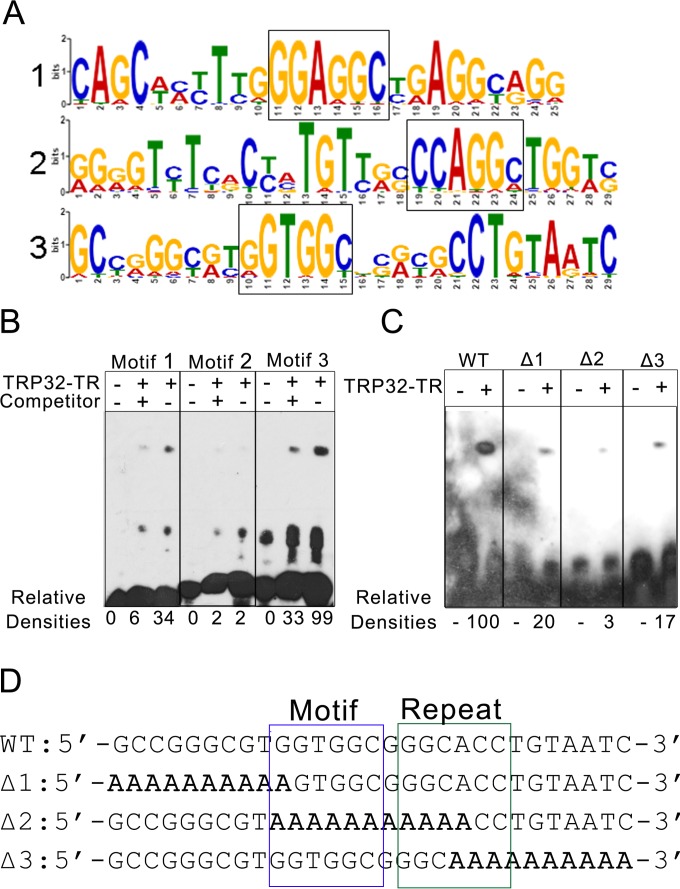
E. chaffeensis TRP32 binds a G-rich motif. (A) TRP32 was predicted to bind to multiple, highly similar, G-rich motifs. Probability matrices of the top three predicted TRP32 DNA motifs from MEME-ChIP are shown. (B) The predicted motifs from panel A were tested by EMSA. TRP32-TR was incubated with biotin-labeled probes in the presence and absence of unlabeled specific competitor. Relative densities are indicated. Motif 3 was identified as the putative DNA motif due to its strong binding, which was significantly decreased (P = 0.002) in the presence of competitor. (C) The TRP32-TR construct was incubated with the wild-type motif 3 (WT) and with three mutant probes. TRP32 bound the wild-type probe with greater affinity than the mutants. All of the mutants showed decreased binding. Relative densities are indicated. (D) Sequences of the wild-type (WT) and mutant probes are shown with the putative motif boxed in blue and the inverted repeat boxed in green. Mutated sequences are bolded.