Fig. 12.
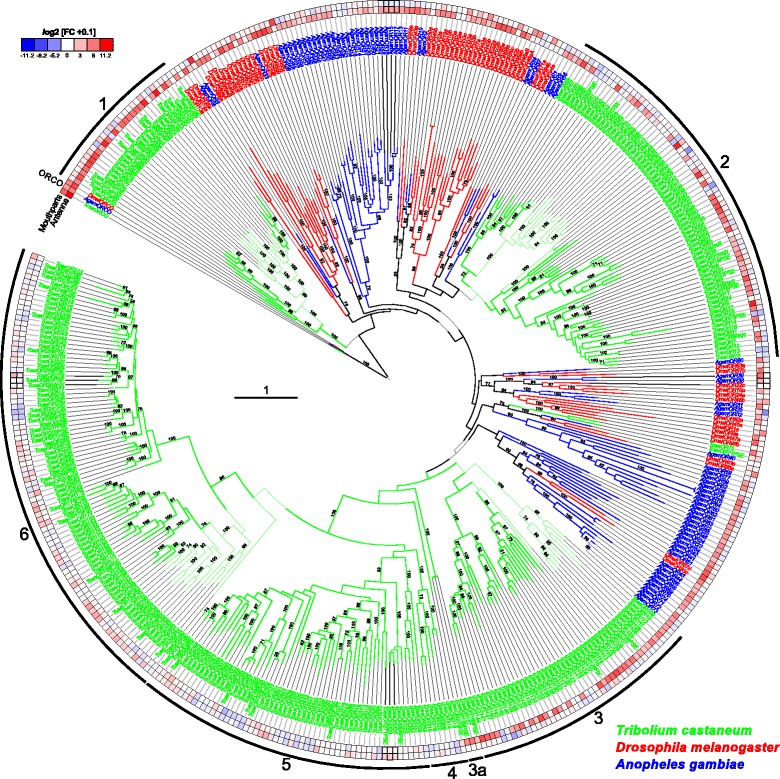
Phylogenetic tree of odorant receptors (ORs). Protein sequences (>300 amino acids) from T. castaneum (green branches), D. melanogaster (red branches), and An. gambiae (blue branches). The tree was rooted using the Orco clade, according to [24]. Robustness of the tree topology was evaluated by 100 rapid bootstrap replications. Outer rings represent the expression in antennae and mouthparts (T. castaneum: palps, mandible, labrum, and labium; D. melanogaster: palp and proboscis; An. gambiae: maxillary palp) as log2-fold change compared to body corresponding to the scale in the left upper corner. The surrounding numbers on the outer thin line indicate the expansion groups 1 to 6 [115]. TcasOR71 and TcasOR72PSE were previously assigned to expansion group 1. The scale bar within the tree represents one amino acid substitution per site. Basically the same figure is available with absolute values instead of fold changes to get an impression of the tissue-specific abundance of the transcripts as Additional file 14: Figure S10. OR odorant receptor
