Fig. 13.
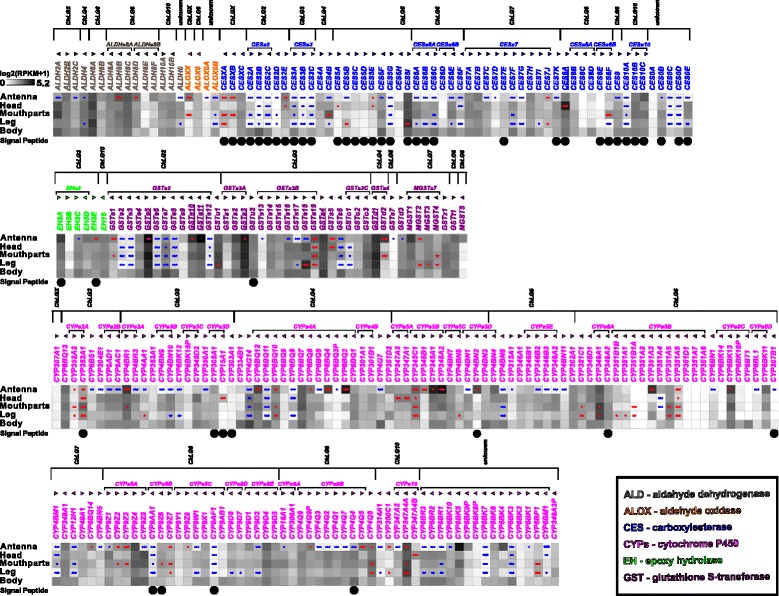
Expression of T. castaneum potential odorant degrading enzymes (ODEs). Heat map showing the expression level of the 263 potential ODEs as a log2[RPKM + 1] value in different tissues [adult antennae, head (missing antennae but including mouthparts), mouthparts, legs, and body]. The candidates are ordered according to their protein family and chromosomal localization. Horizontal brackets above indicate clustering in the genome (Additional file 15: Figure S11), and the arrowheads represent the orientation of the open reading frame. Underlined genes were previously found on the protein level in antennae by [89]. The expression levels are represented by a greyscale with the highest shown expression levels labeled black. The asterisks mark statistically significantly differentially expressed genes compared to body (based on biological replicates of five antennal, two head, three mouthpart, two leg, and two body samples). The red asterisks represent up- and the blue down-regulation (p values adjusted are * < 0.05, ** < 0.01, and *** < 0.001). A black dot in the lowest line indicates a predicted signal peptide according to a SignalP 4.0 [211] prediction. ODE odorant degrading enzyme, RPKM reads per kilobase per million
