Abstract
Dysfunctional telomeres and DNA damage repair (DDR) play important roles in cancer progression. Studies have reported correlations between these factors and tumour aggressiveness and clinical outcome in breast cancer. We studied the characteristics of telomeres and expression of ERCC1, a protein involved in a number of DNA repair pathways and in telomere homeostasis, to assess their prognostic value, alone or in combination, in 90 residual breast tumours after treatment with neoadjuvant chemotherapy (NCT). ERCC1 status was investigated at different molecular levels (protein and gene expression and gene copy‐number variations) by immunohistochemistry, qRT‐PCR and quantitative multiplex fluorescent‐PCR (QMF‐PCR). A comprehensive analysis of telomere characteristics was performed using qPCR for telomere length and qRT‐PCR for telomerase (hTERT), tankyrase 1 (TNKS) and shelterin complex (TRF1, TRF2, POT1, TPP1, RAP1 and TIN2) gene expression. Short telomeres, high hTERT and TNKS expression and low ERCC1 protein expression were independently associated with worse survival outcome. Interestingly, ERCC1 gains and losses correlated with worse disease‐free (p = 0.026) and overall (p = 0.043) survival as compared to survival of patients with normal gene copy‐numbers. Unsupervised hierarchical clustering of all ERCC1 and telomere parameters identified four subgroups with distinct prognosis. In particular, a cluster combining low ERCC1, ERCC1 gene alterations, dysfunctional telomeres and high hTERT and a cluster with high TNKS and shelterin expression correlated with poor disease‐free (HR= 5.41, p= 0.0044) and overall survival (HR= 6.01, p= 0.0023) irrespective of tumour stage and grade. This comprehensive study demonstrates that telomere dysfunction and DDR can contribute synergistically to tumour progression and chemoresistance. These parameters are predictors of clinical outcome in breast cancer patients treated with NCT and could be useful clinically as prognostic biomarkers to tailor adjuvant chemotherapy post‐NCT.
Keywords: Telomeres, telomerase, tankyrase, shelterin, breast cancer, neoadjuvant chemotherapy, ERCC1, prognosis, residual disease
Abbreviations
- POT1
protection of telomere 1
- RAP1
Repressor/Activator Protein 1
- TIN2
TRF1‐ and TRF2‐Interacting Nuclear Protein 2
- TPP1
TIN2 and POT1 interacting protein
- TRF1
telomeric repeat‐binding factor 1
- TRF2
telomeric repeat‐binding factor 2
Introduction
Neoadjuvant chemotherapy (NCT) has been used in breast cancer to facilitate conserving surgery and allows early evaluation of response 1. Patients achieving pathological complete response (pCR) have better disease‐free survival 2. Some characteristics of post‐NCT tumours, such as tumour size or grade, are closely associated with the outcome of patients without pCR 3, 4. These criteria, however, may be misleading. Hence, studying biological prognostic markers for patients without pCR is crucial. Here, we chose to combine DNA damage repair (DDR) and telomeric parameters to assess prognosis in post‐NCT breast cancer patients.
The ERCC1/XPF (excision repair cross complementation group 1/xeroderma pigmentosum factor) heterodimer is a structure‐specific endonuclease mainly known for its involvement in nucleotide excision repair (NER) and a number of other DDR pathways 5. ERCC1 regulates DNA and protein interactions and XPF provides the endonuclease activity 6. ERCC1 has been extensively studied as a putative biomarker of response to treatments 7. Immunohistochemical studies of ERCC1 status conducted in breast cancer patients showed correlations between high expression and good prognosis factors 8. The exact mechanisms underlying this phenomenon remain unclear.
Telomeres are nucleoprotein structures that protect chromosomal ends from exonucleolytic degradation and inappropriate activation of DDR pathways. The shelterin complex comprises six core proteins (TRF1, TRF2, RAP1, TIN2, TPP1 and POT1) and has crucial functions in telomere protection and length regulation. Human telomerase reverse transcriptase (hTERT), a component of telomerase, increases the length of the telomere by adding telomeric repeats to the end of chromosomes 9. Tankyrase 1 (TNKS) is a poly‐ADP‐ribose polymerase that assembles chains of poly‐ADP ribose on the target substrate, particularly TRF1. TNKS is a positive regulator of telomere length by removing TRF1 from telomeres and allowing access to telomerase 10. Telomere shortening due to incomplete DNA synthesis occurs at initial steps of oncogenesis and leads to genomic instability. In contrast, hTERT increases during tumour progression, preventing cancer cells with genomic instability from entering senescence or apoptosis 11. Telomere shortening and telomerase activation have been studied in breast cancer and were correlated with poor prognosis. However, their clinical significance in post‐NCT residual tumours is unknown. In addition, the significance of changes in shelterin and tankyrase gene expression is under‐investigated.
ERCC1‐XPF is also involved in telomere maintenance. Mice null for ERCC1 have a single telomere phenotype characterized by telomeric DNA‐containing double minute chromosomes 12. In cell lines, the overexpression of XPF induces telomere shortening, suggesting that XPF could function as a negative regulator of telomere length via both TRF2‐dependent and independent mechanisms 12, 13. In mouse keratinocytes, XPF mediates telomere degradation induced by TRF2 overexpression 14. Finally, a partial loss of telomeres in TRF2 deficient cells was dependent on ERCC1/XPF expression 12. Interactions between ERCC1 and telomeres have not been studied in the context of tumour cells.
In this study, ERCC1 status and telomeric parameters were investigated at different molecular levels and correlated with clinicopathological data in a cohort of invasive breast cancer patients treated with NCT without pCR. We suggest that post‐NCT tumours with low ERCC1 expression and dysfunctional telomeres are of poor prognosis, whereas high ERCC1 and functional telomeres are good prognostic markers.
Patients and methods
Patients and samples
We retrieved archived tumour samples from 90 women diagnosed with invasive breast cancer and treated with NCT protocols 15 between 1996 and 2010 at the Jean Perrin Cancer Center, Clermont‐Ferrand, France. Clinicopathological data are summarized in supplementary material, Table S1. After NCT, patients underwent appropriate surgery, and radiotherapy. Patients with important residual disease received adjuvant chemotherapy. Trastuzumab was used for the adjuvant treatment and the treatment of recurrent metastatic disease in HER2+ cases. Menopausal patients with hormone receptor‐positive tumours received tamoxifen for 5 years. During follow‐up (median of 9 years), 34 patients (38.9%) relapsed locally and/or presented contralateral or distant events, and 26 (28.9%) died from breast cancer.
Protein studies were performed on paraffin‐embedded tissue sections of 90 post‐NCT tumours and 59 matched pre‐NCT biopsies. Cryopreserved post‐NCT samples were available for molecular investigations for 45 out of 90 patients (Biological Resources Center BB‐0033‐00075). The institutional review board approved the study.
ERCC1 immunohistochemistry (IHC)
A control haematoxylin and eosin‐stained slide was analyzed for each tissue block. ERCC1 IHC staining was operated on a Benchmark ULTRA IHC/ISH staining module (Ventana, USA) as described in Supplementary methods. Semi‐quantitative assessment of ERCC1 expression was performed by two independent readers (FPL, PR) according to the following algorithm: staining intensity on tumour cells was graded on a scale from 0 to 3, with score 3 representing high intensity and endothelial cells in tonsil control tissue used as a reference and assigned an intensity of 2. The percentage of positive tumour cells calculated for each specimen was multiplied by the staining intensity to obtain an H‐score ranging from 0 to 300. Only nuclear staining was scored, but the readers recorded unexpected patterns (eg cytoplasmic). Discordant cases were reviewed. Entire sections were assessed in both pre‐ and post‐NCT samples.
DNA and RNA extraction
DNA and RNA were extracted from snap‐frozen post‐NCT tumour samples with the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Courtaboeuf, France) according to the manufacturer's instructions.
ERCC1 gene copy‐number quantification
The ERCC1 gene copy number was quantified by quantitative multiplex fluorescent polymerase chain reaction (QMF‐PCR). Primers were designed for three fragments of the ERCC1 gene (5′UTR, Exon 6 and 3′UTR). Seven other genes (PVRL1, BOD1L, RET, ZNF638, AGBL2, CFTR and POR) were amplified as controls. QMF‐PCR was performed according to published protocols 16, 17. Primers and technical details are given in Supplementary methods. Fluorescence intensities of PCR products were correlated with the copy number of the relevant exons. Fourteen control DNAs were included in each experiment: 11 normal DNAs and 3 DNAs with a known ERCC1 copy number (deletion, normal and gain). A dosage quotient was calculated relative to all the other amplified exons in patients and controls. The mean ratios ±3 standard deviations of controls were used as cut‐off points to detect copy number variations (CNVs): ratios > 1.2 were considered as gains and ratios < 0.8 as losses.
Telomere length assessment
Average telomere length was measured by quantitative real‐time DNA‐PCR as described elsewhere 18. This method, developed by Cawthon 19, measures the ratio between the telomere repeat (T) and reference single‐copy gene (S) template amounts (relative T/S ratio). We previously showed that the T/S ratio in tumour cells was proportional to the average telomere length assessed by a classical telomere restriction fragment analysis 18.
Gene expression analysis
The gene expression of hTERT and shelterin (TRF1, TRF2, POT1, TPP1, RAP1 and TIN2) was quantified by real‐time RT‐PCR as described previously 18, 20. TNKS (tankyrase 1) and ERCC1 expression was quantified with known primers 21, 22. The normalized copy number (NCN) for each gene was expressed as the ratio between transcript copy‐numbers of the target and control (B2M) genes multiplied by 100.
Statistical analysis
Standard tests (Kruskal‐Wallis test, ANOVA, Student's t‐test, chi‐squared test, Spearman correlation) were used to study the relationship between characteristics. As is usual in exploratory studies, we elected not to adjust probabilities by the Bonferroni method, which, while decreasing the rate of false positives, also increases in a similar proportion the rate of false negatives. Disease‐free survival (DFS) refers to the period between diagnosis and the first relapse (local or distant). Overall survival (OS) was measured from the start of NCT until the last follow‐up report. Survival curves were established by the Kaplan‐Meier method and compared with the log‐rank test. An unsupervised hierarchical clustering was performed with SEM software 23. Distances between clusters were calculated by 1‐Pearson's correlation coefficient values and the dendrogram was constructed according to Ward's algorithm.
Results
ERCC1 protein and gene expression level
In post‐NCT tumours, the median H‐score was 102 (0–285). Positive ERCC1 staining was nuclear (Figure 1A,B). Two patients had negative staining and three cytoplasmic staining. Correlations between clinicopathological data and ERCC1 H‐scores in post‐NCT tumours are shown in Table 1. There was no association between ERCC1 expression and tumour size, grade, stage, hormone‐receptor expression and molecular classification. Interestingly, patients treated with anthracyclines had higher H‐scores than patients receiving anthracyclines + taxanes or taxanes alone (p = 0.004). Comparison of ERCC1 expression in pre‐NCT and post‐NCT samples (ΔERCC1) indicated an increase in ERCC1 levels after treatment with anthracyclines as compared to other regimes (p = 0.042), suggesting that anthracyclines induced ERCC1 expression (Figure 1C,D). After NCT, Chevallier's class 4 patients had lower H‐scores than class 3 patients (p = 0.029). There was a reduction in ERCC1 expression in class 4 patients (ΔERCC1= −72.4) but an increase in class 3 patients (ΔERCC1 = 14.66, p = 0.010). The reduction in ERCC1 levels was correlated with larger residual tumour (p = 0.01). Increased ERCC1 expression after treatment was seen in pre‐NCT Scarff‐Bloom‐Richardson (SBR) grade I tumours (ΔERCC1 = 95.5), but not in grade II‐III tumours, which had stable or decreased ERCC1 levels (grade II=−17.88; grade III=3.2, p = 0.044).
Figure 1.
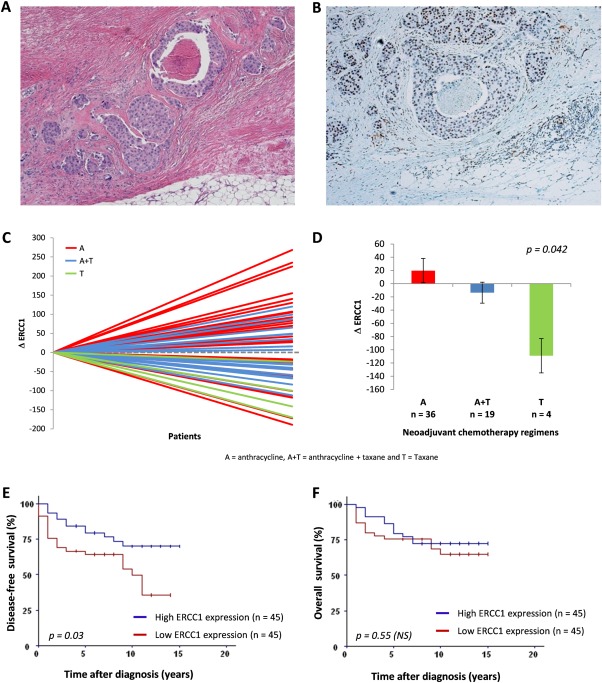
Detection of pre‐ and post‐NCT ERCC1 expression and associations with NCT regimen and survival. (A–B): Representative images of ERCC1‐positive invasive carcinoma: HE staining (A) and corresponding IHC staining (B), showing nuclear positivity in tumour cells. (C‐D): Variations in ERCC1 expression between pre‐NCT and post‐NCT samples (C) are associated with NCT regimens: tumours treated with anthracyclines had an increase only in ERCC1 levels as compared to anthracyclines + taxanes and taxanes groups (D). (E‐F): Survival analysis. Patients whose tumours had low ERCC1 expression (< median, 102) had significantly shortened DFS, whereas those with high ERCC1 levels had better prognosis (E). No significant difference in OS was observed between the groups with low and high ERCC1 levels (F).
Table 1.
ERCC1 protein expression in association with pre‐ or post‐NCT clinical and pathological features of breast cancer patients*
| ERCC1 H score | ||
|---|---|---|
| Variable | Mean (±SEM) | p‐value |
| Tumour size | 0.08 | |
| T1–T2 (n = 66) | 115.9 (±10.5) | |
| T3–T4 (n=24) | 78.9 (±13.1) | |
| Nodal status | 0.53 | |
| Negative (n = 46) | 102.1 (±12.5) | |
| Positive (n = 44) | 110.1 (±11.8) | |
| AJCC clinical stage | 0.53 | |
| I‐II (n = 78) | 108.3 (±9.4) | |
| III (n = 12) | 91.5 (±20.6) | |
| Tumour grade (SBR) | 0.19 | |
| I (n = 9) | 160.4 (±32) | |
| II (n = 46) | 101.1 (±10.7) | |
| III (n = 22) | 101.2 (±16.9) | |
| HR status | 0.34 | |
| Negative (n = 27) | 93.2 (±15.6) | |
| Positive (n = 59) | 110.8 (±10.8) | |
| Molecular classification | 0.57 | |
| Luminal A (n = 23) | 104 (±16.9) | |
| Luminal B (n = 25) | 114 (±16.4) | |
| HER2 (n = 4) | 84 (±37.5) | |
| Triple negative (n = 13) | 77 (±20.2) | |
| Neoadjuvant chemotherapy | 0.0042 | |
| Anthracycline based (n = 47) | 131.2 (±12.7) | |
| Anthracycline and Taxane based (n = 35) | 85.4 (±10.3) | |
| Taxane based (n = 8) | 48.5 (±26.5) | |
| Chevallier's pathological response | 0.029 | |
| Class 3 (n = 70) | 117.6 (±14.1) | |
| Class 4 (n = 14) | 72.6 (±22.3) | |
| Tumour grade (SBR) in residual tumour | 0.19 | |
| I (n = 13) | 131.4 (±24.7) | |
| II (n = 36) | 109.6 (±10.1) | |
| III (n = 19) | 82.2 (±19.8) | |
| HR status in residual tumour | 0.28 | |
| Negative (n = 25) | 97.4 (±15.9) | |
| Positive (n = 60) | 117.8 (±10.4) | |
* Missing values are due to block exhaustion.
AJCC, American Joint Committee on Cancer; SBR, Scarff‐Bloom‐Richardson grading system; HR, hormone receptors; SEM: standard error of the mean.
To evaluate the prognostic impact of ERCC1 expression in residual tumours, we compared survival in patients with low and high H‐scores, using the median value as a cut‐off. Patients with low ERCC1 levels had shorter DFS (p = 0.03). There was no association between ERCC1 expression and OS (Figure 1E,F). Of note, pre‐NCT ERCC1 and ΔERCC1 were not correlated with DFS and OS.
The levels of ERCC1 gene and protein expression evaluated in post‐NCT tumour samples were not correlated. ERCC1 gene expression was not associated with clinicopathological characteristics (data not shown).
ERCC1 CNVs
Of the 45 post‐NCT tumour samples, six had losses and five had gains (Figure 2A–C). These alterations involved the entire ERCC1 gene. In addition, two tumours had partial gains involving the 5′UTR primer only.
Figure 2.
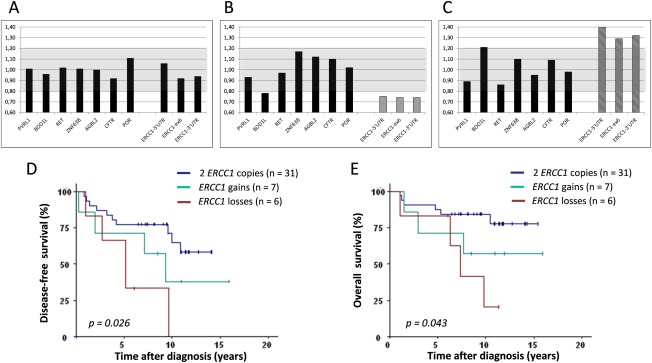
Post‐NCT ERCC1 copy‐numbers: detection and correlation with survival. (A‐C): Determination of ERCC1 gene copy numbers. ERCC1 copy numbers were evaluated by QMF‐PCR with three distinct probes on the ERCC1 gene (localized in 5′UTR, Exon 6 and 3′UTR) and seven other genes serving as controls (PVRL1, BOD1L, RET, ZNF638, AGBL2, CFTR and POR). The gray zone corresponds to normal gene‐copy number, which was defined as the mean ±3 standard deviations of controls. Three examples of ERCC1 profiles are presented: two copies (A), loss (B) and gain (C). (D‐E) Survival analysis. ERCC1 losses and gains were associated with significantly shorter DFS (D) and OS (E) than in the group without CNVs. ERCC1 losses are associated with the worst survival and gains with an intermediate survival.
Patients with gains showed an average increase in ERCC1 gene (75%) and protein (40%) expression, whereas in those with losses ERCC1 gene and protein expression decreased by 20%. However, these changes were not significant.
We compared survival according to ERCC1 gene status: (i) losses, (ii) gains (partial or total) and (iii) normal copy‐number (Figure 2D,E), and found significant differences for DFS and OS. In particular, the subgroup with ERCC1 losses had lower DFS (5.1 years, p = 0.0065) and OS (7.2 years, p = 0.014) than the normal copy‐number subgroup (median not reached). The subgroup with ERCC1 gains had intermediate OS and DFS.
Telomere length and hTERT expression
The median value of hTERT expression was 3.5 (0.0–147.6) and that of telomere length 1.03 (0.22–2.71). Of note, shorter telomeres were associated with higher hTERT expression (p = 0.00075). Correlations between telomere length and hTERT expression with clinicopathological data are summarized in Table 2. Telomere shortening was associated with advanced disease stage (p = 0.030), positive nodal status (p = 0.031) and higher tumour grades in pre‐ and post‐NCT tumours (p = 0.0057 and p = 0.05). hTERT expression tended to increase in advanced stages and higher post‐NCT tumour grades and correlated with hormone‐receptor (HR) negativity in post‐NCT tumours (p = 0.039). No association was observed between telomere parameters and molecular classification. However, patients with triple negative breast cancer (TNBC) tended to have shorter telomeres and higher hTERT. Telomeres were shorter and hTERT higher in cases with the largest residual disease (Chevallier's class 4 versus class 3, p < 0.05). The impact of telomere length and hTERT level on outcome was analyzed using the median values as cut‐points (Figure 3A,B). Patients with short telomeres had lower DFS (p = 0.0076) and OS (p = 0.050). High hTERT expression was also related to decreased DFS (p = 0.0015) and OS (p = 0.025).
Table 2.
Telomere length, hTERT and TNKS expression in association with pre‐ or post‐NCT clinical and pathological features of breast cancer patients*
| Telomere length | hTERT expression | TNKS expression | ||||
|---|---|---|---|---|---|---|
| Variable | Mean (±SEM) | p‐value | Mean (±SEM) | p‐value | Mean (±SEM) | p‐value |
| Tumour size | 0.84 | 0.91 | 0.42 | |||
| T1–T2 (n = 30) | 1.080 (±0.087) | 15.69 (±6) | 345.9 (±49.4) | |||
| T3–T4 (n = 15) | 1.049 (±0.130) | 12.55 (±5.28) | 389.5 (±86.1) | |||
| Nodal status | 0.031 | 0.32 | 0.18 | |||
| Negative (n = 21) | 1.181 (±0.087) | 7.87 (±2.79) | 409 (±70.7) | |||
| Positive (n = 24) | 0.972 (±0.108) | 20.58 (±7.62) | 317.9 (±52.6) | |||
| AJCC clinical stage | 0.030 | 0.10 | 0.68 | |||
| I‐II (n = 38) | 1.128 (±0.078) | 10.53 (±3.38) | 367.8 (±51.3) | |||
| III (n = 7) | 0.751 (±0.135) | 36.99 (±19.2) | 320.3 (±40.9) | |||
| Tumour grade (SBR) | 0.0057 | 0.20 | 0.026 | |||
| I (n = 5) | 1.744 (±0.337) | 1.68 (±19.56) | 326.4 (±147.2) | |||
| II (n = 22) | 0.943 (±0.082) | 17.67 (±5.87) | 303.3 (±54.9) | |||
| III (n = 10) | 1.107 (±0.113) | 23.28 (±0.67) | 540.4 (±116.3) | |||
| HR status | 0.079 | 0.086 | 0.38 | |||
| Negative (n = 11) | 0.841 (±0.102) | 23.82 (±9.9) | 438.9 (±116.8) | |||
| Positive (n = 31) | 1.161 (±0.093) | 11.47 (±5.06) | 335.6 (±47.1) | |||
| Molecular classification | 0.078 | 0.23 | 0.60 | |||
| Luminal A (n = 17) | 1.17 (±0.122) | 8.68 (±4.14) | 343 (±63.6) | |||
| Luminal B (n = 15) | 1.264 (±0.137) | 5.39 (±1.56) | 321.1 (±70.5) | |||
| HER2 (n = 3) | 1.135 (±0.155) | 13.03 (±7.98) | 411.7 (±139.8) | |||
| Triple negative (n = 5) | 0.653 (±0.136) | 36.36 (±18.98) | 531 (±229.5) | |||
| Neoadjuvant chemotherapy | 0.20 | 0.59 | 0.80 | |||
| Anthracycline based (n = 23) | 0.987 (±0.093) | 19.85 (±7.75) | 339.9 (±48.8) | |||
| Anthracycline and Taxane based (n = 22) | 1.155 (±0.108) | 9.20 (±3.43) | 381.9 (±73.4) | |||
| Chevallier's pathological response | 0.045 | 0.041 | 0.50 | |||
| Class 3 (n = 38) | 1.115 (±0.083) | 12.07 (±3.77) | 363.1 (±51) | |||
| Class 4 (n = 5) | 0.740 (±0.041) | 37.12 (±24.76) | 358.2 (±69.7) | |||
| Tumour grade (SBR) in residual tumour | 0.05 | 0.027 | 0.20 | |||
| I (n = 8) | 1.454 (±0.264) | 10.34 (±8.13) | 340.4 (±92.9) | |||
| II (n = 17) | 0.973 (±0.091) | 12.07 (±6.56) | 328.9 (±68.6) | |||
| III (n = 10) | 0.928 (±0.083) | 32.22 (±13.22) | 496 (±122.7) | |||
| HR status in residual tumour | 0.15 | 0.039 | 0.54 | |||
| Negative (n = 12) | 0.869 (±0.098) | 24.81 (±9.21) | 343.3 (±46.8) | |||
| Positive (n = 32) | 1.149 (±0.091) | 10.9 (±4.92) | 370.4 (±58.9) | |||
* Missing values are due to block exhaustion.
AJCC, American Joint Committee on Cancer; SBR, Scarff‐Bloom‐Richardson grading system; HR, hormone receptors; SEM, standard error of the mean.
Figure 3.
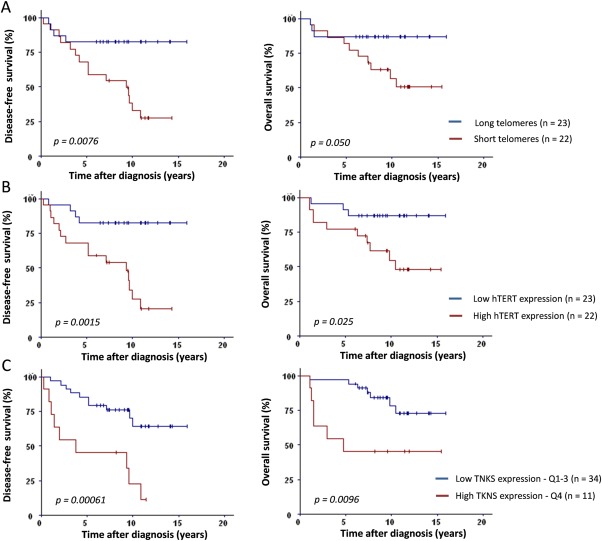
Post‐NCT telomere length, hTERT and TNKS expression levels are predictive of disease‐free and overall survival. (A) Median telomere length (T/S ratio of 1.03) was used to separate tumours into two groups: long (blue line) versus short (red line) telomeres. Short telomeres were significantly associated with worse DFS (p = 0.0076) and OS (p = 0.050). (B) Tumours were divided into low hTERT (blue line) versus high hTERT (red line) expression according to the median value (NCN of 3.5). High hTERT expression was significantly correlated with worse DFS (p = 0.0015) and OS (p = 0.025). (C) Higher TNKS expression (fourth quartile, TNKS ≥ 443, red line) was significantly associated with worse DFS (p = 0.00061) and OS (p = 0.0096).
Shelterin and tankyrase 1 gene expression levels
Shelterin gene expression did not correlate with disease stage, tumour size, nodal status, SBR Grade (pre‐ and post‐NCT), HR status (pre and post‐NCT), pathological response, NCT regimen or survival (data not shown). High expression of the TNKS gene was observed in high SBR grade pre‐NCT tumours (p = 0.026). Higher tankyrase expression (fourth quartile, TNKS > 442) was strongly associated with worse DFS (p = 0.00061) and OS (p = 0.0096) (Figure 3C). Prognostic impact of TNKS was independent of hTERT expression and telomere length (multivariate Cox analysis, adjusted p = 0.013 and p = 0.05, respectively).
Clustering analysis of ERCC1 and telomere parameters
We performed an unsupervised hierarchical clustering analysis to identify prognostic subgroups among 45 patients, combining telomeric and ERCC1 data in post‐NCT tumours. Four balanced groups with different prognosis were defined on the basis of their molecular similarities (Figure 4A–C).
Figure 4.
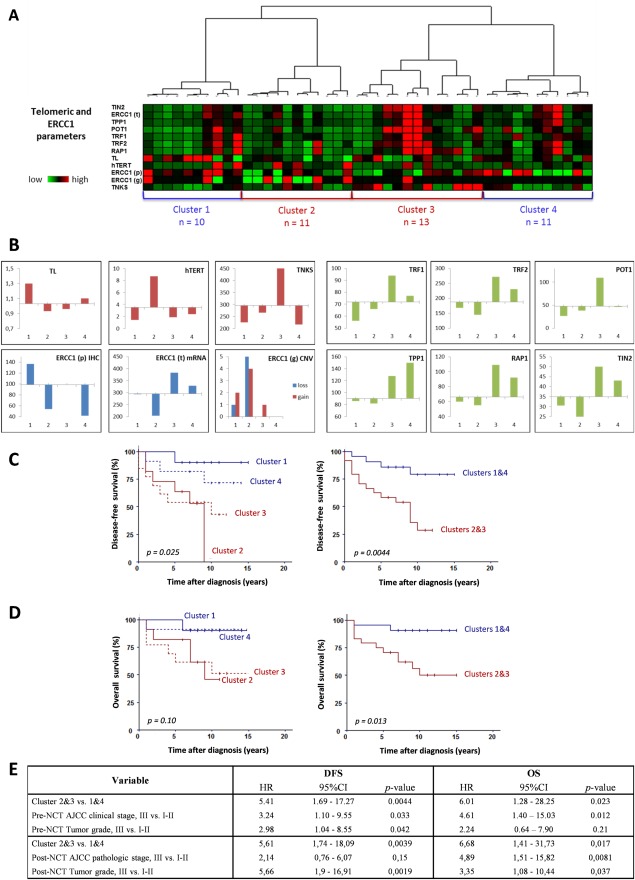
Post‐NCT telomere and ERCC1 characteristics, combined together, correlate statistically with disease‐free and overall survival. (A) Unsupervised hierarchical clustering was performed on the combined telomere length (TL), hTERT, TNKS, shelterin (TRF1, TRF2, TPP1, POT1, RAP1 and TIN2) and ERCC1 gene expression, ERCC1 protein expression and ERCC1 gene CNV. The three levels of analysis for ERCC1 are annotated (p) for protein expression, (t) for gene expression and (g) for copy number. The different color intensities of dichotomic parameters reflect normalized values and not original binary (0/1) values. High normalized quantitative values of telomere length and expression of ERCC1 and telomere‐related genes are shown in red, intermediate values in black and lower values in green (intensity scale is shown). (B) Distribution of the different parameters among the four clusters. (C) Patients from Clusters 2 and 3 (red lines) had shorter DFS than those from clusters 1 and 4 (blue lines) (p = 0.025). A highly significant difference was observed between clusters 2 and 3 versus clusters 1and 4. (D) Clusters 2 and 3 (red lines) tended to be associated with worse OS than clusters 1 and 4 (blue lines). Patients from grouped clusters 1and 4 had significantly worse OS than patients from clusters 2 and 3 (p = 0.013). (E) Prognostic value of these clusters in a multivariate Cox‐model with tumour stage and tumour grade. Patients from clusters 2 and 3 had significantly shorter DFS and OS, irrespective of tumour pre‐ and post‐NCT stage or grade.
Cluster 1 (n = 10) had good prognostic parameters (long telomere, no or very low hTERT expression) and high ERCC1 protein expression levels. Shelterin, TNKS and ERCC1 gene expression levels were low. The ERCC1 gene had few CNVs. These patients had good DFS and OS (9.5 and 9.6 years, means).
Cluster 2 (n = 11) had bad prognostic parameters (short telomere, high expression of hTERT), low ERCC1 levels, low shelterin, TNKS and ERCC1 gene expression. Cluster 2 patients had the highest level of ERCC1 CNVs (both losses and gains) and the lowest DFS and OS (6.2 and 7.8 years, respectively).
In cluster 3 (n = 13), the telomeres were short but hTERT expression low. Shelterin and ERCC1 gene expression levels were the highest. Of note, these patients also had TNKS overexpression. There were a few ERCC1 CNVs, and ERCC1 expression was intermediate. Survival was short in this subgroup (DFS of 6.7 and OS of 8 years).
Cluster 4 (n = 11) had ‘normal’ telomeric parameters, moderate to high shelterin gene expression (in particular TPP1 overexpression) and low TNKS expression levels. The level of ERCC1 gene expression was moderate, that of ERCC1 protein expression low, and no ERCC1 CNVs were observed. Survival data were rather favorable in this cluster (DFS of 9 and OS of 9.5 years).
Survival analysis showed that patients from clusters 1 and 4 had better DFS and OS than patients from clusters 2 and 3 (Figure 4D). Furthermore, patients from clusters 2 and 3 had early relapses with DFS at 5 years of 62.5% compared to DFS at 5 years of 90.5% in clusters 1 and 4 (p = 0.014). A multivariate Cox‐model analysis with pre‐ and post‐NCT tumour stages and grades showed that clusters 2 and 3 correlated with shorter DFS and OS irrespective of tumour stage or grade (Figure 4E). Prognostic profiles of clusters were significant when adjusted for pathological response with hazard ratios of 4.64 for DFS (p = 0.0072) and 5.81 for OS (p = 0.023).
Discussion
There is emerging evidence that pathobiological features of post‐NCT disease can determine outcome in breast cancer 24. The adjuvant treatment decision‐making process, currently based on pre‐NCT biopsy criteria, is beginning to switch toward the use of post‐NCT disease factors. In this study, we investigated ERCC1 expression at different molecular levels and telomere characteristics in post‐NCT breast tumours and correlated the results with patient outcome.
Associations between ERCC1 protein expression and clinicopathological data have been previously explored in breast cancer. Despite their heterogeneous design, most studies suggested that high ERCC1 expression was correlated with criteria of good prognosis, such as estrogen‐receptor positivity and low T/N stages 8, 25. Low ERCC1 levels were found in poor‐prognosis TNBC 26, 27. Our results show better survival in cases of high ERCC1 in post‐NCT tumours, but not in pre‐NCT tumours. We may infer that ERCC1 expression in residual tumours is a robust prognostic marker in breast cancer patients treated with NCT.
NCT protocols influence DDR mechanisms and pathological response. pCR rate increases after platinum‐based CT in tumours underexpressing ERCC1 28. This is in line with the paradigm that negative/low ERCC1 tumours respond better to platinum‐based CT because of insufficient NER activation 29, 30. In addition, anthracycline‐based CT may induce an increase in ERCC1 gene expression levels in breast tumours because of the activation of ERCC1‐dependent DDR mechanisms other than NER 31. Our study showed increasing ERCC1 expression in tumours treated with anthracyclines compared with tumours treated with taxanes. Post‐NCT ERCC1 status could be taken into account in future tailoring of platinum‐based adjuvant therapy.
ERCC1 expression was greater in post‐NCT Chevallier class 3 tumours than in class 4 cases. Better response is generally observed in tumours with higher genomic instability 32 while hormone‐receptor positive tumours, which have low genomic instability, are usually poor responders to NCT 33. Class 3 and 4 patients represent heterogeneous groups in terms of genomic instability and other biological characteristics. It is possible that ERCC1 expression levels could help to identify better responders among HR‐positive cancer patients like those with luminal B tumours, which have higher levels of genomic instability and are more chemosensitive than luminal A tumours 34.
In agreement with other reports, we did not observe any prognostic significance of ERCC1 mRNA expression at the gene level 35. In addition, copy‐number alterations of ERCC1 gene were previously shown to be highly frequent in non‐small cell lung cancer but not correlated with protein expression and survival 36. We found that ERCC1 CNVs tended to be associated with parallel variations in protein and gene expression levels and that both gains and losses were correlated with worse outcome. We suggest that ERCC1 protein functionality could be affected by CNVs.
In naïve breast cancer, telomere shortening was associated with tumour aggressiveness according to TNM stage or SBR grade 22, 37, 38, 39. Short telomeres were found in aggressive HER2+ and TNBC cases 40, 41. In our series of post‐NCT tumours, we also observed telomere shortening in patients with advanced stage and grade.
A relationship between short telomeres and poor clinical outcomes has already been reported 37, 42, 43 but not confirmed in a study with longer median follow‐up time (7.2 years) 44. Furthermore, the impact of short telomeres on DFS was recently observed in TNBC but not in HR‐positive tumours 45. Our analysis of a cohort with a long follow‐up (9 years) and a majority of HR‐positive tumours strongly supports the overall link between short telomeres and poor survival in breast cancer. In order to maintain short telomeres, hTERT is activated in tumour cells, and high hTERT correlates with clinical aggressiveness in many human malignancies 18, 46, 47, 48. In agreement with previous findings in breast cancer patients 22, 49, 50, we observed that increased hTERT expression correlated with higher SBR grades and shorter survival. Thus, we suggest that telomere length and hTERT expression in post‐NCT tumours are significant predictors of survival and could be used to tailor adjuvant chemotherapy.
Overexpression of TNKS has been described in breast cancer in comparison to normal tissue 51 and has been associated with higher SBR grades 22. We found that TNKS overexpression appeared mainly in high‐grade SBR III tumours, suggesting that this overexpression represents a relevant event in tumour progression. We also demonstrated that high TNKS expression was associated with shorter survival. Because of its involvement in telomere maintenance, tankyrase was recently considered as a potential target for cancer therapy 52, 53. The correlation of TNKS expression with survival underlines the validity of this target.
Down‐regulation of shelterin genes TRF1, TRF2 and POT1 has been found in breast cancer compared with normal breast tissue 22, 50, 54, 55. Low expression of POT1, TRF1 and TRF2 tended to be associated with high tumour grade and poor clinical outcome, but this relationship was not significant 22, 50. We observed no correlation between shelterin gene expression and clinicopathological parameters or survival.
Clustering analysis combining telomeric and ERCC1 parameters identified four subgroups with different prognosis and with possibly different biological significance. In particular, the analysis of poor‐prognosis cluster 2 patients showed that telomere‐dysfunction parameters (short telomere length, high hTERT, low shelterin) were associated with decreased ERCC1 expression and ERCC1 copy‐number alterations suggestive of a certain deficiency in DDR mechanisms. Both telomere and DDR defects can contribute to a higher degree of genomic instability and, given the possible involvement of ERCC1 in telomere maintenance, amplify the deleterious impact of telomere dysfunction, resulting in resistant tumour phenotypes. The hTERT overexpression observed in this cluster could have contributed to tumour cell proliferation by stabilizing short telomeres and by conferring survival advantages owing to the telomere‐independent function of hTERT as transcriptional modulator of the NF‐ΚB and Wnt/ß‐catenin pathways 56.
Another poor‐prognosis association, including high TNKS, ERCC1 and shelterin gene expression, was found in cluster 3 patients. Telomere length and hTERT were less affected, but this subgroup had worse prognosis than clusters 1 and 4 with no marked telomere changes. TNKS acts as a positive regulator of telomere elongation in telomerase‐positive cells and also has telomere‐independent functions that contribute to correct bipolar spindle assembly during mitosis 57 and the maintenance of Wnt‐signalling 58. All these functions of tankyrase may increase the survival capacities of tumour cells. In addition, up‐regulation of ERCC1 and shelterin genes may have an adverse effect on telomeres. Shelterin interacts with proteins involved in DDR, such as ERCC1‐XPF. Overexpression of XPF may negatively regulate telomere length maintenance and interfere with shelterin binding to telomeres 13.
The association of long telomeres, low hTERT expression, low shelterin and high ERCC1 protein expression that we observed in cluster 1 patients, irrespective of tumour stage or grade, confirms that together functional telomere and DDR protein recruitment determine the best prognosis.
In conclusion, we report new findings on the clinical significance of ERCC1, a DDR protein, and telomere parameters in breast cancer treated with NCT. The combination of these characteristics identifies subgroups with different prognosis. These results emphasize the interest in studying DDR proteins and their role in telomere maintenance in translational studies.
Author Contributions
MGB and PR performed experiments, generated and analyzed data and wrote the manuscript draft. AC, LV, MP, SS, PC, FK, CA, YJB and PV contributed to experiments, data collection and revision of the manuscript for content, analysis and interpretation of data. AT and FPL designed the study and wrote the final manuscript.
Supporting information
SUPPLEMENTARY MATERIAL ONLINE
Supplementary methods
Table S1. Clinico‐pathological characteristics of breast cancer patients included in this study
Acknowledgements
This work was supported in part by grants from the French National Cancer Institute and the League Against Cancer. PR was recipient of a scholarship from Roche Ventana in Tucson AZ, USA. Ventana provided the ERCC1 Antibody. Esteban Roberts, Staff Scientist at Roche Ventana in Tucson AZ, USA, contributed to the elaboration of the ERCC1 IHC procedure.
Conflict of interest statement: The authors declare no conflict of interest.
References
- 1. Buchholz TA, Hunt KK, Whitman GJ, et al Neoadjuvant chemotherapy for breast carcinoma: multidisciplinary considerations of benefits and risks. Cancer 2003; 98: 1150–1160. [DOI] [PubMed] [Google Scholar]
- 2. Fisher B, Bryant J, Wolmark N, et al Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J Clin Oncol 1998; 16: 2672–2685. [DOI] [PubMed] [Google Scholar]
- 3. Penault‐Llorca F, Abrial C, Raoelfils I, et al Changes and predictive and prognostic value of the mitotic index, Ki‐67, cyclin D1, and cyclo‐oxygenase‐2 in 710 operable breast cancer patients treated with neoadjuvant chemotherapy. Oncologist 2008; 13: 1235–1245. [DOI] [PubMed] [Google Scholar]
- 4. Pierga JY, Mouret E, Dieras V, et al Prognostic value of persistent node involvement after neoadjuvant chemotherapy in patients with operable breast cancer. Br J Cancer 2000; 83: 1480–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kirschner K, Melton DW. Multiple roles of the ERCC1‐XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res 2010; 30: 3223–3232. [PubMed] [Google Scholar]
- 6. Tripsianes K, Folkers G, Ab E, et al The structure of the human ERCC1/XPF interaction domains reveals a complementary role for the two proteins in nucleotide excision repair. Structure 2005; 13: 1849–1858. [DOI] [PubMed] [Google Scholar]
- 7. McNeil EM, Melton DW. DNA repair endonuclease ERCC1‐XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res 2012; 40: 9990–10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Goyal S, Parikh RR, Green C, et al Clinicopathologic significance of excision repair cross‐complementation 1 expression in patients treated with breast‐conserving surgery and radiation therapy. Int J Radiat Oncol Biol Phys 2010; 76: 679–684. [DOI] [PubMed] [Google Scholar]
- 9. Diotti R, Loayza D. Shelterin complex and associated factors at human telomeres. Nucleus 2011; 2: 119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol 2000; 10: 1299–1302. [DOI] [PubMed] [Google Scholar]
- 11. Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis 2010; 31: 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu XD, Niedernhofer L, Kuster B, et al ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA‐containing double minute chromosomes. Mol Cell 2003; 12: 1489–1498. [DOI] [PubMed] [Google Scholar]
- 13. Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech Ageing Dev 2008; 129: 602–610. [DOI] [PubMed] [Google Scholar]
- 14. Munoz P, Blanco R, Flores JM, et al XPF nuclease‐dependent telomere loss and increased DNA damage in mice overexpressing TRF2 result in premature aging and cancer. Nat Genet 2005; 37: 1063–1071. [DOI] [PubMed] [Google Scholar]
- 15. Penault‐Llorca F, Abrial C, Raoelfils I, et al Comparison of the prognostic significance of Chevallier and Sataloff's pathologic classifications after neoadjuvant chemotherapy of operable breast cancer. Hum Pathol 2008; 39: 1221–1228. [DOI] [PubMed] [Google Scholar]
- 16. Niel F, Martin J, Dastot‐Le Moal F, et al Rapid detection of CFTR gene rearrangements impacts on genetic counselling in cystic fibrosis. J Med Genet 2004; 41: e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pebrel‐Richard C, Kemeny S, Gouas L, et al An atypical 0.8 Mb inherited duplication of 22q11.2 associated with psychomotor impairment. Eur J Med Genet 2012; 55: 650–655. [DOI] [PubMed] [Google Scholar]
- 18. Veronese L, Tournilhac O, Callanan M, et al Telomeres and chromosomal instability in chronic lymphocytic leukemia. Leukemia 2013; 27: 490–493. [DOI] [PubMed] [Google Scholar]
- 19. Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res 2002; 30: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Poncet D, Belleville A, t'kint de Roodenbeke C, et al Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B‐chronic lymphocytic leukemia. Blood 2008; 111: 2388–2391. [DOI] [PubMed] [Google Scholar]
- 21. Ganzinelli M, Mariani P, Cattaneo D, et al Expression of DNA repair genes in ovarian cancer samples: biological and clinical considerations. Eur J Cancer 2011; 47: 1086–1094. [DOI] [PubMed] [Google Scholar]
- 22. Poonepalli A, Banerjee B, Ramnarayanan K, et al Telomere‐mediated genomic instability and the clinico‐pathological parameters in breast cancer. Genes Chromosomes Cancer 2008; 47: 1098–1109. [DOI] [PubMed] [Google Scholar]
- 23. Kwiatkowski F, Girard M, Hacene K, et al [Sem: a suitable statistical software adaptated for research in oncology]. Bull Cancer 2000; 87: 715–721. [PubMed] [Google Scholar]
- 24. Bossuyt V, Provenzano E, Symmans WF, et al Recommendations for standardized pathological characterization of residual disease for neoadjuvant clinical trials of breast cancer by the BIG‐NABCG collaboration. Ann Oncol 2015; 26: 1280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerhard R, Carvalho A, Carneiro V, et al Clinicopathological significance of ERCC1 expression in breast cancer. Pathol Res Pract 2013; 209: 331–336. [DOI] [PubMed] [Google Scholar]
- 26. Kim D, Jung W, Koo JS. The expression of ERCC1, RRM1, and BRCA1 in breast cancer according to the immunohistochemical phenotypes. J Korean Med Sci 2011; 26: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sidoni A, Cartaginese F, Colozza M, et al ERCC1 expression in triple negative breast carcinoma: the paradox revisited. Breast Cancer Res Treat 2008; 111: 569–570. [DOI] [PubMed] [Google Scholar]
- 28. Chen X, Wu J, Lu H, et al Measuring beta‐tubulin III, Bcl‐2, and ERCC1 improves pathological complete remission predictive accuracy in breast cancer. Cancer Sci 2012; 103: 262–268. [DOI] [PubMed] [Google Scholar]
- 29. Lee KB, Parker RJ, Bohr V, et al Cisplatin sensitivity/resistance in UV repair‐deficient Chinese hamster ovary cells of complementation groups 1 and 3. Carcinogenesis 1993; 14: 2177–2180. [DOI] [PubMed] [Google Scholar]
- 30. Fu JM, Zhou J, Shi J, et al Emodin affects ERCC1 expression in breast cancer cells. J Transl Med 2012; 10 Suppl 1: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fu JM, Zhou J, Xie JS, et al [Effect of neoadjuvant chemotherapy on ERCC1 gene expression in breast cancer]. Nan Fang Yi Ke Da Xue Xue Bao 2008; 28: 603–605. [PubMed] [Google Scholar]
- 32. Ribeiro E, Ganzinelli M, Andreis D, et al Triple negative breast cancers have a reduced expression of DNA repair genes. PLoS One 2013; 8: e66243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Colleoni M, Viale G, Zahrieh D, et al Chemotherapy is more effective in patients with breast cancer not expressing steroid hormone receptors: a study of preoperative treatment. Clin Cancer Res 2004; 10: 6622–6628. [DOI] [PubMed] [Google Scholar]
- 34. Ades F, Zardavas D, Bozovic‐Spasojevic I, et al Luminal B breast cancer: molecular characterization, clinical management, and future perspectives. J Clin Oncol 2014; 32: 2794–2803. [DOI] [PubMed] [Google Scholar]
- 35. Deng Q, Yang H, Lin Y, et al Prognostic value of ERCC1 mRNA expression in non‐small cell lung cancer, breast cancer, and gastric cancer in patients from Southern China. Int J Clin Exp Pathol 2014; 7: 8312–8321. [PMC free article] [PubMed] [Google Scholar]
- 36. Vanhecke E, Valent A, Tang X, et al 19q13‐ERCC1 gene copy number increase in non–small‐cell lung cancer. Clin Lung Cancer 2013; 14: 549–557. [DOI] [PubMed] [Google Scholar]
- 37. Fordyce CA, Heaphy CM, Bisoffi M, et al Telomere content correlates with stage and prognosis in breast cancer. Breast Cancer Res Treat 2006; 99: 193–202. [DOI] [PubMed] [Google Scholar]
- 38. Kammori M, Sugishita Y, Okamoto T, et al Telomere shortening in breast cancer correlates with the pathological features of tumor progression. Oncol Rep 2015; 34: 627–632. [DOI] [PubMed] [Google Scholar]
- 39. Martinez‐Delgado B, Gallardo M, Tanic M, et al Short telomeres are frequent in hereditary breast tumors and are associated with high tumor grade. Breast Cancer Res Treat 2013; 141: 231–242. [DOI] [PubMed] [Google Scholar]
- 40. Heaphy CM, Asch‐Kendrick R, Argani P, et al Telomere length alterations unique to invasive lobular carcinoma. Hum Pathol 2015; 46: 1197–1203. [DOI] [PubMed] [Google Scholar]
- 41. Heaphy CM, Subhawong AP, Gross AL, et al Shorter telomeres in luminal B, HER‐2 and triple‐negative breast cancer subtypes. Mod Pathol 2011; 24: 194–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heaphy CM, Baumgartner KB, Bisoffi M, et al Telomere DNA content predicts breast cancer‐free survival interval. Clin Cancer Res 2007; 13: 7037–7043. [DOI] [PubMed] [Google Scholar]
- 43. Simpson K, Jones RE, Grimstead JW, et al Telomere fusion threshold identifies a poor prognostic subset of breast cancer patients. Mol Oncol 2015; 9: 1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lu L, Zhang C, Zhu G, et al Telomerase expression and telomere length in breast cancer and their associations with adjuvant treatment and disease outcome. Breast Cancer Res 2011; 13: R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nagelkerke A, van Kuijk SJ, Martens JW, et al Poor prognosis of constitutive gamma‐H2AX expressing triple‐negative breast cancers is associated with telomere length. Biomark Med 2015; 9: 383–390. [DOI] [PubMed] [Google Scholar]
- 46. Kirkpatrick KL, Mokbel K. The significance of human telomerase reverse transcriptase (hTERT) in cancer. Eur J Surg Oncol 2001; 27: 754–760. [DOI] [PubMed] [Google Scholar]
- 47. Taubert H, Wurl P, Greither T, et al Stem cell‐associated genes are extremely poor prognostic factors for soft‐tissue sarcoma patients. Oncogene 2007; 26: 7170–7174. [DOI] [PubMed] [Google Scholar]
- 48. Uen YH, Lin SR, Wu DC, et al Prognostic significance of multiple molecular markers for patients with stage II colorectal cancer undergoing curative resection. Ann Surg 2007; 246: 1040–1046. [DOI] [PubMed] [Google Scholar]
- 49. Bieche I, Nogues C, Paradis V, et al Quantitation of hTERT gene expression in sporadic breast tumors with a real‐time reverse transcription‐polymerase chain reaction assay. Clin Cancer Res 2000; 6: 452–459. [PubMed] [Google Scholar]
- 50. Salhab M, Jiang WG, Newbold RF, et al The expression of gene transcripts of telomere‐associated genes in human breast cancer: correlation with clinico‐pathological parameters and clinical outcome. Breast Cancer Res Treat 2008; 109: 35–46. [DOI] [PubMed] [Google Scholar]
- 51. Gelmini S, Poggesi M, Distante V, et al Tankyrase, a positive regulator of telomere elongation, is over expressed in human breast cancer. Cancer Lett 2004; 216: 81–87. [DOI] [PubMed] [Google Scholar]
- 52. Riffell JL, Lord CJ, Ashworth A. Tankyrase‐targeted therapeutics: expanding opportunities in the PARP family. Nat Rev Drug Discov 2012; 11: 923–936. [DOI] [PubMed] [Google Scholar]
- 53. Seimiya H. The telomeric PARP, tankyrases, as targets for cancer therapy. Br J Cancer 2006; 94: 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Diehl MC, Idowu MO, Kimmelshue KN, et al Elevated TRF2 in advanced breast cancers with short telomeres. Breast Cancer Res Treat 2011; 127: 623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saito K, Yagihashi A, Nasu S, et al Gene expression for suppressors of telomerase activity (telomeric‐repeat binding factors) in breast cancer. Jpn J Cancer Res 2002; 93: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y, Tergaonkar V. Noncanonical functions of telomerase: implications in telomerase‐targeted cancer therapies. Cancer Res 2014; 74: 1639–1644. [DOI] [PubMed] [Google Scholar]
- 57. Palazzo L, Della Monica R, Visconti R, et al ATM controls proper mitotic spindle structure. Cell Cycle 2014; 13: 1091–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bao R, Christova T, Song S, et al Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PLoS One 2012; 7: e48670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL ONLINE
Supplementary methods
Table S1. Clinico‐pathological characteristics of breast cancer patients included in this study


