Abstract
Innate lymphoid cells (ILCs) are innate immune cells that are ubiquitously distributed in lymphoid and nonlymphoid tissues and enriched at mucosal and barrier surfaces. Three major ILC subsets are recognized in mice and humans. Each of these subsets interacts with innate and adaptive immune cells and integrates cues from the epithelium, the microbiota, and pathogens to regulate inflammation, immunity, tissue repair, and metabolic homeostasis. Although intense study has elucidated many aspects of ILC development, phenotype, and function, numerous challenges remain in the field of ILC biology. In particular, recent work has highlighted key new questions regarding how these cells communicate with their environment and other cell types during health and disease. This review summarizes new findings in this rapidly developing field that showcase the critical role ILCs play in directing immune responses through their ability to interact with a variety of hematopoietic and nonhematopoietic cells. In addition, we define remaining challenges and emerging questions facing the field. Finally, this review discusses the potential application of basic studies of ILC biology to the development of new treatments for human patients with inflammatory and infectious diseases in which ILCs play a role.
Introduction
The innate lymphoid cell (ILC) family consists of a variety of innate immune cells that lack antigen specificity, are enriched at mucosal and barrier surfaces, and regulate immune responses and tissue homeostasis (Mebius, 2003; Tait Wojno and Artis, 2012; Spits et al., 2013; Vosshenrich and Di Santo, 2013; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Classical NK cells were the first ILCs identified, and they promote protective immune responses against pathogens and tumor cells (Vosshenrich and Di Santo, 2013). Lymphoid tissue inducer (LTi) cells that orchestrate the formation of secondary lymphoid tissues were subsequently identified and later classified as members of the ILC family (Mebius, 2003). More recently, groundbreaking work in murine models and numerous studies of human tissue samples has expanded the family to include the group 1, 2, and 3 ILC subsets (ILC1s, ILC2s, and ILC3s, respectively), which are defined by the cytokines and transcription factors they express and their effector functions (Spits et al., 2013). These cells produce cytokines, integrate environmental signals, and interact with other immune cells to regulate immunity, inflammation, and tissue homeostasis (Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Although all ILCs arise from a common precursor early in development, elegant studies have revealed that classical NK cell development branches off and is distinct from that of other ILC subsets and that classical NK cells perform unique functions compared with other ILCs (Vosshenrich and Di Santo, 2013). Therefore, this review solely focuses on the noncytolytic, “helper-like” ILCs.
Noncytolytic ILCs arise from a common lymphoid progenitor found in the bone marrow (Diefenbach et al., 2014; Artis and Spits, 2015). The development of these cells either requires or involves the γ chain (γc) cytokine IL-7; the transcription factors inhibitor of DNA binding 2 (Id2; Eberl et al., 2004; Moro et al., 2010; Satoh-Takayama et al., 2010; Cherrier et al., 2012; Klose et al., 2014), GATA-binding protein 3 (GATA3; Serafini et al., 2014; Yagi et al., 2014), promyelocytic leukemia zinc finger protein (PLZF; Constantinides et al., 2014), nuclear factor interleukin-3 regulated (Nfil3; Geiger et al., 2014; Seillet et al., 2014), T cell factor-1 (TCF1; Mielke et al., 2013; Yang et al., 2015a), and thymocyte selection–associated high-mobility group box (TOX; Seehus et al., 2015); and Flt3 ligand (Baerenwaldt et al., 2016). ILCs populate lymphoid organs but are particularly enriched at mucosal and barrier surfaces and are rapid and potent cytokine producers that participate in a variety of immune responses (Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Indeed, the noncytolytic, helper-like ILC1s, ILC2s, and ILC3s parallel the major CD4+ Th type 1, 2, and 17 (Th1, Th2, and Th17) subsets and express similar transcription factors and cytokines (Spits et al., 2013; Robinette et al., 2015). ILC1s express the transcription factor T-box expressed in T cells (T-bet) and produce IFN-γ, making them key contributors to cell-mediated immune responses to bacteria and protozoan parasites (Sciumé et al., 2012; Bernink et al., 2013; Fuchs et al., 2013; Klose et al., 2014). ILC2s express GATA3, type 2 cytokines such as IL-4, IL-5, IL-9, and IL-13, the growth factor amphiregulin (Areg), and met-enkephalin peptides to promote type 2 immune responses that protect against helminth infection, cause allergic inflammation, mediate tissue repair, and maintain metabolic homeostasis (Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Monticelli et al., 2011; Kim et al., 2013b; Odegaard and Chawla, 2013; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015; Brestoff et al., 2015). Finally, ILC3s express retinoid-related orphan receptor γt (RORγt) and produce IL-17A, IL-22, and lymphotoxin (Mebius et al., 1997; Satoh-Takayama et al., 2008; Sawa et al., 2010; Sonnenberg et al., 2011b, 2012; McKenzie et al., 2014). ILC3s are the most heterogeneous ILC population and can be divided into three main groups: LTi cells and LTi-like C-C chemokine receptor type 6 (CCR6)–expressing ILC3s that contribute to the development of lymphoid structures and intestinal inflammation and the T-bet+ natural cytotoxicity receptor NCR+ and NCR− ILC3s that promote tissue homeostasis, antibacterial immunity, and autoimmune inflammation (Fig. 1, inset; Mebius et al., 1997; Satoh-Takayama et al., 2008; Sawa et al., 2010; Sonnenberg et al., 2011b, 2012; Klose et al., 2013; Rankin et al., 2013; McKenzie et al., 2014).
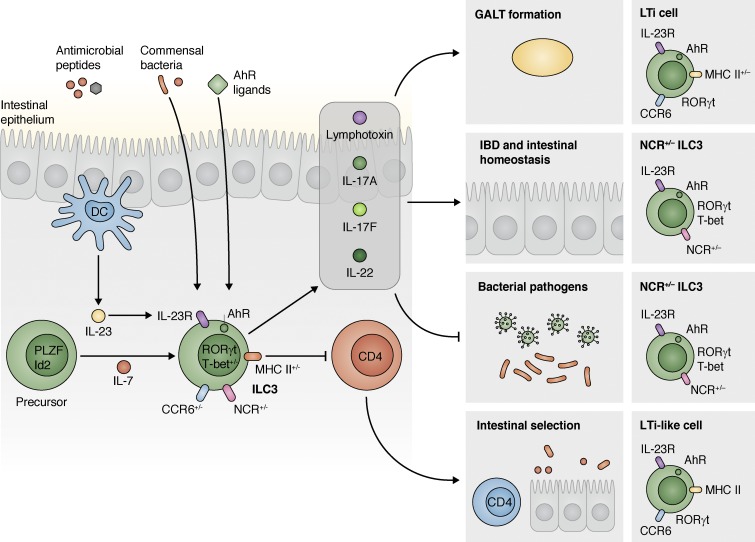
Figure 1.
ILC3s promote GALT formation, inflammation, immunity, and homeostasis in the intestine. A progenitor that expresses multiple transcription factors, including PLZF and Id2, responds to IL-7 and differentiates into RORγt-expressing ILC3s (though notably CCR6-expressing/LTi cell progenitors do not express PLZF). Mature ILC3s then integrate multiple environmental cues that modulate their effector functions. These cells are activated by IL-23 produced by myeloid cells through expression of the IL-23R. They can also respond to signals from the microbiota and dietary factors, such as AhR ligands and vitamins. Together, these signals regulate the ability of ILC3s to produce cytokines such as lymphotoxin, IL-17, and IL-22. Three major ILC3 subsets exist, including the CCR6+NCR−T-bet− LTi and LTi-like cells, the CCR6−NCR−T-bet+ ILC3s, and the CCR6−NCR+T-bet+ ILC3s. ILC3-derived cytokines then promote GALT formation (LTi cells), regulate intestinal homeostasis and inflammation during IBD and in other contexts (CCR6−NCR+/−T-bet+ ILC3s), and contribute to protective immunity to intestinal pathogens (CCR6−NCR+/−T-bet+ ILC3s). Finally, LTi-like ILC3s express MHC class II, which allows them to interact with and delete commensal-specific CD4+ T cells to maintain tolerance of commensal bacterial species and intestinal homeostasis.
Despite the parallels between ILC and Th subsets, ILCs lack expression of antigen-specific receptors and do not directly mediate antigen-specific responses (Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Instead, ILCs coordinate signals from the epithelium, the microbiota, pathogens, and other immune cells by expressing an array of cytokine and eicosanoid receptors and cytokines, thus acting as central organizers of immune responses (Eberl, 2012; Pearson et al., 2012; Kim et al., 2013b; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). The development of tools for ILC analysis has led to an explosion of studies unveiling new aspects of ILC development and function. This review will focus on recent work that highlights the unique abilities of ILCs to interact with their environment and other immune cells to orchestrate complex and diverse biological processes. In addition, we will highlight new challenges and questions that have emerged in the field. Finally, we will explore one of the major challenges in the study of ILCs: to develop therapies that selectively target ILC activities to treat human diseases.
ILC3s interact with the microbiota and innate and adaptive immune cells to regulate intestinal immune responses
LTi cells were the first non-NK cell members of the ILC family to be identified (Mebius et al., 1997; Mebius, 2003; Pearson et al., 2012). Later work characterized similar cells, including CCR6-expressing LTi-like ILC3s and NCR-expressing ILC3s, that participate in the formation of gut-associated lymphoid tissues (GALTs), including cryptopatches and isolated lymphoid follicles (ILFs), maintenance of intestinal homeostasis, and protection against infection (Fig. 1; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Recent studies have begun to unravel how ILC3s interface with the microbiota and various cell types to maintain intestinal function. New findings have led to questions related to how ILC3s are regulated throughout life, how they sense bacteria- and diet-derived factors to promote tolerance of commensal bacteria, and how they regulate intestinal immunity.
ILC3 hematopoiesis and lymphoid tissue formation
Mebius et al. (1997) first described murine CD4+CD3− LTi cells that promoted proper formation of secondary lymphoid tissues. A related LTi-like population was identified in the cryptopatches of mice (Kanamori et al., 1996), and these cells were shown to interact with ILF B cells to promote IgA production (Tsuji et al., 2008). In particular, expression of lymphotoxin by ILC3s is critical for their ability to support IgA production and lymphoid tissue development (Tsuji et al., 2008; Kruglov et al., 2013). Numerous studies have since described factors that regulate the development and function of these cells. Seminal studies from Littman and colleagues and others demonstrated that RORγt expression is required for the development of murine LTi cell and LTi-like ILC3s (Eberl et al., 2004; Satoh-Takayama et al., 2008; Sanos et al., 2009; Eberl, 2012). These RORγt-expressing ILC3s were required for formation of cryptopatches, ILFs (Eberl and Littman, 2004; Eberl et al., 2004; Kiss et al., 2011; Lochner et al., 2011; Lee et al., 2012) and Peyer’s patches (Bando et al., 2015) in mice. Importantly, RORγt is also expressed by human ILC3 progenitors (Montaldo et al., 2014), and humans have LTi-like cells (Cupedo et al., 2009), indicating that these cells and their functions are conserved in mice and humans. Other factors, including the aryl hydrocarbon receptor (AhR) and Notch (Kiss et al., 2011; Possot et al., 2011; Cherrier et al., 2012; Lee et al., 2012; Tait Wojno and Artis, 2012; Rankin et al., 2013), runt-related transcription factor 3 (Runx3; Ebihara et al., 2015), and postnatal microbial colonization of the intestine, control the development and size of the murine LTi cell population (Sawa et al., 2010). In addition, recent work from Ishizuka et al. (2016) showed that PLZF and TCF1 expression gradients control the committed development of LTi cells versus that of other ILC3s, suggesting that LTi cells are distinct from other ILC3s, though this remains to be fully elucidated, particularly in humans.
Together, these data identify a network of factors that influence ILC3 development and function. However, many questions remain. For example, whether ILC3 development requires the microbiota is controversial, as some studies have shown that ILC3s that express NKp46 were absent in germ-free mice (Satoh-Takayama et al., 2008; Sanos et al., 2009), whereas other studies have demonstrated that this subset and/or other ILC3 subsets remained intact (Sanos et al., 2009; Sawa et al., 2010, 2011; Reynders et al., 2011; Lee et al., 2012; Sonnenberg et al., 2012). Notably, studies in murine models have shown that GALT formation is largely independent of the microbiota, with the exception of ILF formation (Bouskra et al., 2008), but in contrast, maternal microbial colonization during gestation increases ILC3 population size in pups (Xiong et al., 2016), suggesting a complex relationship between ILC3 development, the microbiota, and GALT formation. In particular, how the microbiota affects the turnover and circulation of ILC3s and maintenance and organization of GALT in adults is unclear, though recent evidence suggests that specific species of commensal bacteria such as segmented filamentous bacteria can promote ILC3 production of IL-22 (Atarashi et al., 2015) and IL-17 (Sano et al., 2015) in adult mice. In addition, whether ILC3s orchestrate organization of tertiary lymphoid structures in tissues outside of the intestine in unknown. NCR-expressing ILC3s have been associated with the formation of tertiary lymphoid structures in the lung of humans with nonsmall cell lung cancer (Carrega et al., 2015), suggesting that diverse ILC3s in mice and humans might control lymphopoiesis throughout life in various tissues. Finally, although ILC-derived lymphotoxin supports communication between ILCs and T cells or DCs (Cupedo et al., 2004; Eberl et al., 2004; Eberl, 2012; Kruglov et al., 2013) and ILC interactions with stromal cells are also prominent (Mebius, 2003), the precise nature of the interactions between ILC3s and other cells that reside in the GALT is not fully defined. Thus, further studies will be required to understand how the pathways that regulate ILC3 development, function, and turnover shape GALT and tertiary lymphoid structure formation. As much of our understanding of these cells has been based on mechanistic studies in murine models, expanded analysis of human LTi cells will be required to fully appreciate the role of this population in humans throughout the lifespan.
ILC3s maintain the health of the superorganism
RORγt+ ILC3s in mice and humans support intestinal homeostasis by responding to IL-23, producing tissue-protective IL-17A, IL-17F, and IL-22, and interacting with various intestinal cell types and the microbiota (Fig. 1; Eberl, 2012; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). In particular, the complex nature of the relationship between ILC3s and the intestinal microbiota has been a topic of great interest. Although there is controversy regarding whetherthe microbiota shapes development of ILC3s, microbial products have been shown to influence levels of intestinal IL-22, a tissue-protective cytokine made predominantly by RORγt-expressing ILC3s in both mice and humans that promotes production of antimicrobial peptides and mucins (Sonnenberg et al., 2011a; Eberl, 2012; Diefenbach et al., 2014; Artis and Spits, 2015). How this process is regulated is also unclear, as murine ILC IL-22 production is not affected after alteration of commensal bacterial communities in some studies (Sonnenberg et al., 2012), whereas other work showed that germ-free mice had decreased IL-22–expressing NKp46+ ILC3s (Satoh-Takayama et al., 2008; Sanos et al., 2009; Vonarbourg et al., 2010), and still other studies showed that the microbiota could repress murine ILC3-derived IL-22 through IL-25 production (Zaph et al., 2008; Sawa et al., 2011).
Although further work will be needed to understand whether the microbiota is required for IL-22 production by ILCs, particularly in humans, the microbiota clearly can affect ILC3 function and the return to homeostasis after inflammation and infection (Fig. 1). In response to intestinal damage and alterations in the microbiota caused by dextran sodium sulfate, murine ILC3s expanded and up-regulated IL-22, associated with a return to homeostasis (Sawa et al., 2011). Interestingly, specific commensal bacterial species appear to be able to differentially regulate ILC3 function, as segmented filamentous bacteria can promote IL-17 and IL-22 production from murine ILC3s (Atarashi et al., 2015; Sano et al., 2015). Notably, viruses can also modulate ILC3 activities (Abt et al., 2016), as murine norovirus engaged TLR7 on DCs in mice, leading to IL-23 production, ILC3 expression of IL-22, and increased resistance to infection with vancomycin-resistant Enterococcus faecium in microbiota-depleted hosts (Abt et al., 2016).
In addition to the role for the microbiota in shaping ILC3 responses, ILC3s in turn affect the composition and localization of commensal bacterial species. For example, IL-22–expressing LTi-like cells were critical for the anatomical containment of lymphoid-resident commensal Alcaligenesspecies and preventing systemic inflammation in mice (Sonnenberg et al., 2012; Fung et al., 2016). Importantly, antibody responses against Alcaligenes species were also associated with inflammatory bowel disease (IBD) in humans, suggesting that ILC3s may help to shape commensal microbial communities in both mice and humans (Sonnenberg et al., 2012). In support of this idea, lymphoid tissue–resident commensal bacteria that reside within murine DCs in the GALT elicited IL-10 production that limited Th17 responses and promoted resistance to inflammation (Fung et al., 2016).
Parallel to the role of ILC3s in maintaining a host-protective relationship with the microbiota, dysregulation of this relationship can be pathogenic, as IL-23 overexpression in mice was associated with inflammatory ILC3 responses that caused intestinal lesions that could be ameliorated with antibiotic treatment (Chen et al., 2015b). In addition, IL-17A– and IFN-γ–expressing ILC3s promoted colitis in mice (Buonocore et al., 2010), and ILC3-like cells that expressed IL-17 were associated with IBD in humans (Geremia et al., 2011). Thus, the relationship between ILC3s and the microbiota must be carefully regulated (Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Of note, ILC3s have also been associated with inflammation outside of the intestine in both mice and particularly humans, such as the skin (Kim et al., 2013b; Teunissen et al., 2014), and systemically during common variable immunodeficiency (Cols et al., 2016), but the nature of the relationship between ILC3s and the microbiota in these tissues is unclear.
Numerous questions remain regarding the mechanisms by which ILC3s communicate with the microbiota and other environmental factors in the intestine and elsewhere. To begin to address this, recent studies have focused on how intestinal ILC3s sense dietary substances to regulate intestinal immunity (Fig. 1) and how ILC3s can also regulate intestinal homeostasis by influencing the “diet” of the microbiota. Vitamin A is sensed by murine ILC3s, and vitamin A deficiency was associated with impaired ILC3 responses (Spencer et al., 2014). In addition, maternal intake of retinoic acid regulated murine LTi cell responses and immune fitness in pups (van de Pavert et al., 2014), and retinoic acid caused murine ILC3s to up-regulate gut-homing receptors (Kim et al., 2015). In addition, vitamin D deficiency was associated with altered commensal bacterial communities and increased ILC3 responses in mice (Chen et al., 2015a). Phytochemicals derived from cruciferous vegetables also stimulate the AhR and promote intestinal ILC3 activities in mice (Kiss et al., 2011), and cholesterol biosynthetic intermediates, ligands for RORγt, can regulate murine LTi cells (Santori et al., 2015).
Parallel with their ability to react to host dietary compounds, ILC3s can also facilitate the transfer of fucose, which commensal bacteria use as a carbohydrate source, to the surface of murine intestinal epithelial cells. This process maintained an appropriate relationship between the intestinal epithelium and the microbiota, which was critical for resistance to infection with Salmonella typhimurium (Goto et al., 2014). However, despite the connections between ILC3s, diet, and the microbiota, little is known regarding how ILC3s sense signals derived directly from the microbiota, as ILC3 responses are largely intact in mice that lack pattern recognition receptors, and ILC3s do not appear to express such receptors (Sawa et al., 2010). Because products derived from bacterial metabolism, such as short-chain fatty acids, indole, ethanolamine, alcohols, lactate, and others, can influence immune cell activities (Lopez et al., 2014), it is possible that ILC3s could respond to these metabolites, but this remains to be fully explored. Importantly, our understanding of how dietary and microbial products affect human ILC3 function in the intestine is very limited, and further studies in diverse populations of healthy humans and those with diseases associated with altered nutrient availability and microbial colonization are needed.
Finally, recent studies have explored how ILC3s and the microbiota affect the activities of other intestinal immune cells. In particular, interactions between ILC3s and myeloid cells are important. For example, the microbiota conditioned macrophages to produce IL-1β, which elicited ILC3 production of granulocyte-macrophage colony-stimulating factor that supported T regulatory cell (T reg cell) function in mice (Mortha et al., 2014). In addition, CX3CR1+ myeloid cells were shown to receive signals from the microbiota to up-regulate ILC3-derived IL-22 to protect against infection with the murine bacterial pathogen Citrobacter rodentium (Longman et al., 2014). New studies have highlighted that ILC3s also shape adaptive immune responses in the intestine. Studies in mouse models by Sonnenberg and colleagues showed that MHC class II–expressing ILC3s interacted with and deleted CD4+ T cells specific for commensal antigens, demonstrating that ILC3s promote tolerance of the microbiota (Fig. 1; Hepworth et al., 2013, 2015). Conversely, ILC3s isolated from the spleen of mice can also present antigens to elicit proinflammatory adaptive responses in the presence of adjuvants (von Burg et al., 2014). Thus, further studies using murine models of tolerance and inflammation, coupled with selective inhibition of pathways that ILC3s use to sense the microbiota in mice and associative studies in humans, will be needed to understand how NCR− and NCR+ ILC3s interact with immune cells to maintain intestinal homeostasis and whether the activities of these cells could be targeted therapeutically to treat inflammatory diseases of the intestine.
ILC3s protect the host during infection and contribute to anticancer responses
ILC3s also take center stage when the host is infected with microbial pathogens (Fig. 1; Eberl, 2012; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). After intestinal infection in mice with the attaching and effacing bacterium C. rodentium, a protective role for ILC3-derived IL-22 was initially described (Satoh-Takayama et al., 2008; Zheng et al., 2008; Sonnenberg et al., 2011b), and subsequent work showed that ILC3 expression of Runx3 played a key role in this process (Ebihara et al., 2015). In the intestine, studies of murine models have shown that ILC3s interface with adaptive immune cells (Withers et al., 2012), innate immune cells, and epithelial cells (Giacomin et al., 2015) during infection. Myeloid cells associated with ILC3s through the CXCL16–CXCR6 pathway after C. rodentium infection of mice, which elicited ILC3-derived IL-22 and resistance to infection (Satoh-Takayama et al., 2014), and in a virus-induced murine gastroenteritis model, IL-22–producing ILC3s were associated with protective IFN-λ responses (Hernández et al., 2015). In addition, expression of inhibitor of κB kinase α or β in intestinal epithelial cells was required for efficient production of IL-22 from ILC3s and protection from C. rodentium in mice (Giacomin et al., 2015). Thus, IL-22–producing ILC3s are critical in mediating protective immune responses against intestinal pathogens (Fig. 1).
Importantly, recent work has shed light on how different ILC3 subsets contribute to intestinal homeostasis and resistance to bacterial pathogens. Studies from Klose et al. (2013) and Rankin et al. (2013) described T-bet–expressing NCR+RORγt+ ILC3s that either promote resistance to or regulate homeostasis in response to bacterial infection in the intestine. CCR6−RORγt+ ILCs up-regulated T-bet in response to signals from IL-23 and the microbiota (Klose et al., 2013), leading to Notch-directed NKp46 expression (Klose et al., 2013; Rankin et al., 2013), consistent with a previous study (Sciumé et al., 2012). Supporting a role for these cells during intestinal bacterial infection, T-bet–deficient mice had decreased NCR+RORγt+ ILC3s and were more susceptible to infection with C. rodentium (Klose et al., 2013; Rankin et al., 2013). T-bet expression also enhanced IFN-γ production in NCR+RORγt+ ILC3s that promoted enterocolitis after Salmonella enterica infection, but in parallel directed the release of glycoproteins that protected the epithelial barrier (Klose et al., 2013), demonstrating that these T-bet–expressing ILC3s can modulate the balance between host protection and intestinal homeostasis (Fig. 1). Interestingly, recent studies suggest that NCR+ ILC3s are not critically important for murine antibacterial immune responses in the presence of wild-type T cells (Song et al., 2015; Rankin et al., 2016), suggesting that effector functions may be compartmentalized in the ILC3 subsets or that redundancy exists to ensure that antibacterial immunity remains intact in the presence or absence of an effective T cell response. Notably, we know little regarding the role of ILC3s in protecting the human host during intestinal bacterial infection, though the recently described transcriptional heterogeneity of human ILC3s, as analyzed using single-cell RNA-sequencing (Björklund et al., 2016), suggests that compartmentalization of ILC3 effector function may also be conserved in humans.
ILC3s are also associated with protective immune responses in tissues other than the intestine (Tait Wojno and Artis, 2012; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Experiments by Aujla et al. (2008) showed that IL-22 mediated resistance to infection with Klebsiella pneumoniae in the murine lung, though the source of IL-22 in this study was presumed to be Th17 cells. A recent study has shown that cross-talk between monocytes and IL-17–producing ILC3s mediates innate immune defense against this antibiotic-resistant pathogen (Abt et al., 2015). ILC3-like cells in the lung also mounted protective responses after vaccination with Bacillus Calmette–Guérin in mice (Pitt et al., 2012), and ILC3s accumulated in the murine lung and produced IL-22 after Streptococcus pneumoniae infection (Van Maele et al., 2014). After infection with lymphocytic choriomeningitis virus in mice, IL-22–producing LTi-like cells helped to restore lymphoid architecture in secondary lymphoid tissues that were required for responses to secondary antigens (Scandella et al., 2008). Finally, in a murine model of oropharyngeal candidiasis, ILC3s in the oral mucosa produced IL-17A and IL-17F and were required for protective immunity (Gladiator et al., 2013).
Finally, ILC3s can be tissue protective in contexts other than infection, particularly cancer and cancer treatment. For example, IL-22–producing LTi cells support thymic regeneration in mice after total-body irradiation (Dudakov et al., 2012), and activated ILC3s in the peripheral blood of human leukemia patients that received a hematopoietic stem cell transplant were associated with reduced susceptibility to graft-versus-host disease (Munneke et al., 2014). Importantly, ILC3s can play a direct role in anticancer responses. ILC3s respond to IL-12 to promote leukocyte invasion of B16 melanoma tumors in mice (Eisenring et al., 2010), and NCR-expressing ILC3s have been found in human small cell lung tumors and are correlated with the density of protective tertiary lymphoid structures (Carrega et al., 2015). In an oncogene-induced murine cancer model, non-NK, non–T innate lymphocytes expanded early after tumorigenesis and limited tumor growth, although these cells resembled ILC1s (Dadi et al., 2016). Evidence also exists that ILC3s can contribute to the development of cancer, as IL-22 produced by ILC3s can support the development of colorectal cancer in murine models, IL-23 overexpression was associated with ILC3 responses that drove intestinal adenoma formation (Chan et al., 2014), and IL-22–producing CD3− cells can be detected in human colorectal cancer (Kirchberger et al., 2013). Ultimately, extensive studies in murine models of cancer and in human cancer patients will be required to fully understand how ILC3s mediate tissue-protective responses and inflammation during cancer by interacting with the microbiota, the epithelium, and immune cells at multiple mucosal and barrier sites.
ILC2s at the nexus of inflammation, tissue repair, and homeostasis
In 2010, three laboratories independently described a murine ILC population that drove type 2 immune responses (Moro et al., 2010; Neill et al., 2010; Price et al., 2010), and the next year two laboratories identified analogous populations in multiple fetal and adult human tissues (Mjösberg et al., 2011; Monticelli et al., 2011). These cells, now referred to as ILC2s, promote protective immunity to helminth infection, allergic inflammation, tissue repair, and metabolic homeostasis (Kim et al., 2013b; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). As has been the case with ILC3s, intensive interest in ILC2s has led to new challenges and questions in the field. In particular, questions remain regarding ILC2 development, how ILC2s interact with epithelial and immune cells, and how they balance their abilities to maintain homeostasis, promote repair, and cause inflammation.
A regulatory network governs ILC2 development and function
Recent work has shown that ILC2 development and function depends on a complex network of pathways. Specific transcription factors, such as RORα (Halim et al., 2012b; Wong et al., 2012), TCF1 (Yang et al., 2013), growth factor independent 1 transcription repressor (Gfi1; Spooner et al., 2013), and B cell CLL/lymphoma 11b (Bcl11b; Walker et al., 2015; Yu et al., 2015), contribute to murine ILC2 development. Some of these factors are also critical for ILC2 effector functions, as continuing expression of GATA3 in mature ILC2s is needed for normal murine ILC2 function (Hoyler et al., 2012), and loss of Bcl11b expression results in loss of ILC2 gene expression and function and acquisition of ILC3-like properties in mice (Califano et al., 2015).
After development, cytokines derived from epithelial cells, including IL-25, IL-33, and thymic stromal lymphopoietin (TSLP), and immune cell–derived γc cytokines, such as IL-2, IL-4, and IL-7, promote murine and human ILC2 survival and activation (Moro et al., 2010; Neill et al., 2010; Price et al., 2010; Kim et al., 2013b, 2014; Diefenbach et al., 2014; McKenzie et al., 2014; Motomura et al., 2014; Oliphant et al., 2014; Artis and Spits, 2015; Roediger et al., 2015). IL-2 derived from T cells (Oliphant et al., 2014) or other ILCs (Roediger et al., 2015) is particularly important. Additional proteins that influence murine ILC2s include IL-9 (Turner et al., 2013; Mohapatra et al., 2016), the TNF family member tumor necrosis factor–like ligand 1A (Meylan et al., 2014; Yu et al., 2014), and the receptor for advanced glycation end products, an immunoglobulin superfamily molecule produced by nonhematopoietic cells in the lung (Oczypok et al., 2015). Interestingly, new work has shown that human ILC2s can also respond to IL-12 signaling to up-regulate IFN-γ and T-bet expression, indicating that human ILC2s have the ability to respond to a diverse set of environmental signals (Lim et al., 2016).
Recent work has also highlighted a role for nonprotein factors, including bioactive lipids such as eicosanoids, in regulating ILC2 functions. Mjösberg et al. (2011) initially identified human ILC2s by expression of the prostaglandin D2 (PGD2) receptor chemoattractant receptor–homologous molecule expressed on Th2 cells (CRTH2). Indeed, many of the studies describing the effect of PGD2 on ILC2s have been in humans, establishing a role for the PGD2–CRTH2 pathway in mediating human ILC2 chemotaxis, cytokine production, and possibly accumulation in the lung (Barnig et al., 2013; Xue et al., 2014; Tait Wojno et al., 2015), with increases in CRTH2+ ILC2s observed in the peripheral blood of allergic asthma patients (Bartemes et al., 2014) and esophageal tissues of patients with the allergic disease eosinophilic esophagitis (Doherty et al., 2015). Murine and human ILC2s respond to other eicosanoids, including leukotriene D4, which enhanced their function (Doherty et al., 2013), lipoxin A4 (LXA4), which limited their activation (Barnig et al., 2013), and PGI2, which inhibited IL-33–elicited IL-5 and IL-13 production in mice (Zhou et al., 2016). Interestingly, little is currently known about how ILC2s interact with mast cells and basophils that produce eicosanoids, though TSLP-responsive ILCs in the inflamed skin of mice and humans interact with both cell types (Roediger et al., 2013; Kim et al., 2014) and IL-4 derived from basophils supported ILC2 responses in a murine model of allergen-induced airway eosinophilia (Motomura et al., 2014). This work suggests that cytokine and bioactive lipid pathways converge to regulate ILC2 responses, but further studies will be required to reveal how the interplay of transcription factors, cytokines, and bioactive lipids coordinate murine and human ILC2 activities.
ILC2s interact with the epithelium and immune cells to promote type 2 inflammation
ILC2s were formally first identified in murine fat-associated lymphoid clusters and during infection with the rodent nematode parasite Nippostrongylus brasiliensis (Moro et al., 2010; Neill et al., 2010; Price et al., 2010). Based on observations that adaptive cells could not fully account for type 2 cytokine production required for helminth expulsion using mouse models of infection (Fallon et al., 2006), seminal studies revealed that innate cells, later designated ILC2s, were the predominant source of IL-13 after N. brasiliensis infection that was required for changes in the intestinal epithelium that lead to parasite expulsion (Fig. 2; Moro et al., 2010; Neill et al., 2010; Price et al., 2010). These studies have been extended to show that ILC2s also promote immune responses to Strongyloides venezuelensis in mice (Yasuda et al., 2012), suggesting that these cells are key mediators of murine antihelminth responses against multiple species. Of note, how these cells contribute to human resistance to helminth parasites remains uncertain. A recent study has shown that children infected with Schistosoma haematobium had decreased peripheral blood ILC2s (Nausch et al., 2015), suggesting that ILC2s may depopulate the blood and accumulate in tissues in helminth-infected individuals. However, more studies are needed to determine whether ILC2s are as critical for helminth resistance in humans as they are in mice.
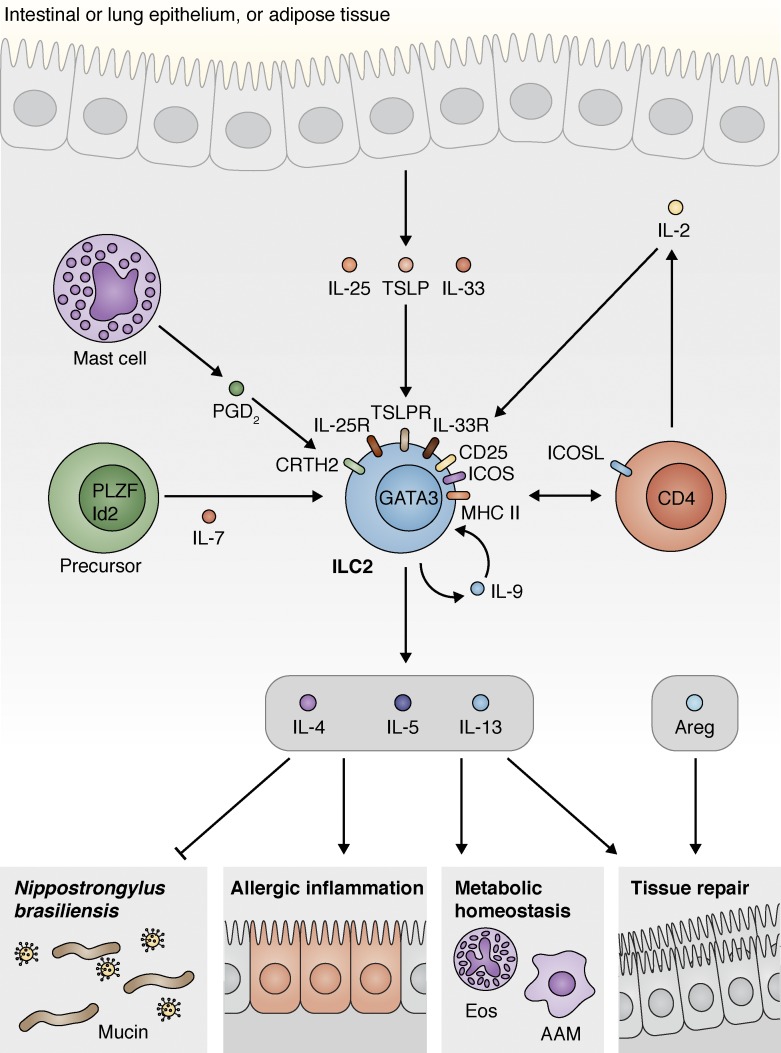
Figure 2.
ILC2s influence inflammation, immunity, tissue repair, and homeostasis through interactions with hematopoietic and nonhematopoietic cells. ILC2s that express GATA3 differentiate from a progenitor cell that expresses a variety of transcription factors, including PLZF and Id2. These progenitors respond to IL-7 and other factors as they differentiate. Once mature, ILC2s sense multiple signals, including cytokines that can be derived from the epithelium, including IL-25, IL-33, and TSLP, and mast cell–derived eicosanoids such as PGD2 and others. Activated ILC2s produce type 2 cytokines, including IL-4, IL-5, IL-9, and IL-13. During N. brasiliensis infection, these cytokines are protective and elicit mucin production that induces helminth expulsion. These cytokines can also cause type 2 inflammation associated with allergic disease in multiple tissues. IL-9 feeds back in an autocrine loop to promote ILC2 survival during inflammation, and IL-4 produced by basophils also supports ILC2 population expansion (not depicted). The proinflammatory effects of ILC2s are balanced by their ability to promote homeostasis and tissue repair. In this context, type 2 cytokines derived from ILC2s support wound healing and maintain eosinophil (Eos) and alternatively activated macrophage (AAM) populations in adipose tissue, which, together with ILC2-derived met-enkephalin peptides, promote and support the lean state. Finally, activated ILC2s can produce growth factors that directly act on the epithelium to repair damaged tissues.
The latest work has again used murine models to dissect how interactions between ILC2s and other cell types coordinate antihelminth responses. Cytokines produced by epithelial cells, such as IL-25 and IL-33, elicit distinct populations of innate cells that contribute to antihelminth immunity in murine models (Saenz et al., 2010, 2013; Huang et al., 2015b). Additionally, a recent study demonstrated that IL-25 drove the emergence of inflammatory ILC2s that expressed killer cell lectin-like receptor G1 (KLRG1), differentiated into classical ILC2s, and supported N. brasiliensis expulsion from the murine intestine (Huang et al., 2015b). Interestingly, these inflammatory ILC2s could also produce IL-17 (Spooner et al., 2013; Huang et al., 2015b) and provided protection against Candida albicans (Huang et al., 2015b), suggesting that murine inflammatory ILC2s can combat an array of pathogens. New studies have identified the Tuft cell, a chemosensory intestinal epithelial cell type, as a primary source of IL-25 in the context of parasite infection in mice (Gerbe et al., 2016; Howitt et al., 2016; von Moltke et al., 2016). In addition to their relationship with epithelial cells, ILC2s also maintain a dialogue with adaptive CD4+ T cells (Fig. 2). Studies by Halim et al. (2014), Mirchandani et al. (2014), and Oliphant et al. (2014) together demonstrated that murine ILC2s express MHC II and interact with CD4+ T cells, supporting Th2 cell differentiation and ILC2 function during helminth infection or allergic disease, with a key role for CD4+ T cell–derived IL-2 in promoting ILC2 responses. In addition, IL-33 and T cell–derived IL-2 caused the ILC2 population to expand in the lung after rechallenge of mice with N. brasiliensis that led to parasite larval damage and prevention of reinfection (Bouchery et al., 2015). Finally, recent work has shown that inducible T cell costimulator (ICOS)–ICOS ligand (ICOSL) interactions between ILC2s and T reg cells led to the development of helminth-associated T reg cell responses in mice (Molofsky et al., 2015).
In addition to their role in helminth-induced type 2 inflammation, ILC2s drive allergic inflammatory diseases, including allergic airway inflammation in mice (Halim et al., 2012a; Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015) and humans (Bartemes et al., 2014; Christianson et al., 2015), and allergic inflammation in the skin and intestine (Kim et al., 2013a,b; Roediger et al., 2013; Salimi et al., 2013). As during helminth infection, pro-allergic ILC2 responses are regulated by interactions with various cell types (Fig. 2; Barnig et al., 2013; Doherty et al., 2013; Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015; Tait Wojno et al., 2015). Nonhematopoietic cells affect ILC2 responses, as structural airway cells that produce IL-33 and TSLP (Mohapatra et al., 2016) and pulmonary epithelial cells that produce transforming growth factor β (Denney et al., 2015) shaped ILC2 responses in murine models of allergic airway inflammation. In similar models, nociceptor neurons in the lung responded to ILC2-derived IL-5 and the neuropeptide vasoactive intestinal peptide to promote allergic inflammation (Gallant et al., 2007). In addition, interactions between human ILC2s that express NKp30 and keratinocytes that express the activating ligand B7-H6 may promote atopic dermatitis (Salimi et al., 2016).
ILC2s also have a complex communication network with other hematopoietic cells in the context of allergic disease. Interactions between ILC2s and CD4+ T cells drive allergic inflammation (Halim et al., 2012a; Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015), and ILCs also interact closely with other innate cells. For example, interactions among ILC2s (Maazi et al., 2015) and between ILCs and DCs (Kamachi et al., 2015) mediated through the ICOS–ICOSL pathway drive allergic inflammation in the murine lung (Montaldo et al., 2014; Kamachi et al., 2015; Maazi et al., 2015). ILC2 interactions with mast cells (Roediger et al., 2013) and basophils (Kim et al., 2014) are associated with atopic dermatitis–like disease in human and murine skin. Recent studies have highlighted an association between human ILC2s and eosinophils during chronic rhinosinusitis (Mjösberg et al., 2011; Kim et al., 2013b; Walford et al., 2014) and eosinophilic esophagitis (Doherty et al., 2015). Finally, type 1 IFN and IL-27 produced by innate cells antagonize ILC2 function to limit allergic lung inflammation (Duerr et al., 2016; Mchedlidze et al., 2016; Moro et al., 2016). Together, these data show that ILC2s communicate with nonhematopoietic and hematopoietic cell types to mediate allergic inflammation, suggesting that these interactions might be targeted to treat ILC2-associated allergic diseases in humans.
Thus, interactions between ILC2s, the epithelium, and other immune cells mediate protective immunity to helminth parasites and allergic inflammation (Fig. 2), but numerous questions remain. Future studies will require the use of sophisticated genetic and imaging tools in murine models of helminth infection and allergy to better understand the molecular mechanisms by which ILC2s interface with the epithelium, other hematopoietic cells, and the microbiota and how ILC2 responses are turned off. An additional challenge is to determine how ILC2s function during human parasitic infection and allergic disease and how these cells regulate the balance between these distinct type 2 inflammatory states. The finding that human S. haematobium infection was associated with a decrease in ILC2s in the peripheral blood (Nausch et al., 2015) could suggest that S. haematobium actively modulates human ILC2 responses. Consistent with this idea, in murine models, Heligmosomoides polygyrus elicited IL-1β that decreased IL-25 and IL-33 levels to support chronicity of infection (Zaiss et al., 2013), and products of H. polygyrus could limit ILC2 responses and allergic lung inflammation (McSorley et al., 2014). These findings provoke the hypothesis that ILC2s influence the relationship between helminth infection and allergy, and future studies investigating ILC2 regulation, particularly in helminth-infected and allergic humans, should provide new insight into how these cells balance protective and pathological inflammation in multiple tissues.
ILC2s limit inflammation and promote tissue homeostasis
In addition to their inflammatory properties, ILC2s play diverse roles in limiting inflammation, regulating tissue repair, and maintaining homeostasis (Fig. 2; Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). One mechanism through which this occurs is via production of type 2 cytokines, which can regulate type 1 or type 17 inflammation (McKenzie et al., 2014; Artis and Spits, 2015). Decreased numbers of type 2 cytokine–expressing ILC2s were associated with experimental murine autoimmune encephalitis (Russi et al., 2015), and type 2 cytokine–expressing ILC2s supported tissue protection in a murine model of renal reperfusion injury (Huang et al., 2015a) and biliary atresia (Li et al., 2014). In addition, cigarette smoke altered IL-33 responsiveness in the murine lung to limit ILC2 production of type 2 cytokines, associated with susceptibility to viral infection (Kearley et al., 2015). Importantly, the ability of ILC2s to participate in tissue repair through type 2 cytokine production can become dysregulated, as pulmonary fibrosis in mice after Schistosoma mansoni infection or Bleomycin administration (Hams et al., 2014) and liver fibrosis (Mchedlidze et al., 2013) was dependent on IL-13–producing ILC2s. Thus, further studies will be required to determine how ILC2s can repair tissues without contributing to fibrosis, particularly in humans.
ILC2s also express factors that directly lead to wound healing, such as the growth factor Areg, which was critical in mediating repair of the damaged respiratory and intestinal epithelium after murine influenza A virus infection and intestinal injury and inflammation, respectively (Monticelli et al., 2011, 2015). Areg-producing ILC2s that responded to IL-9 have also been associated with lung tissue repair after helminth infection in mice (Turner et al., 2013), and IL-33–responsive ILC2s promote cutaneous wound healing in mice and are associated with acute wounds in human skin (Rak et al., 2016). These data suggest that ILC2s contribute to tissue repair using multiple mechanisms, and future studies may reveal that these cells produce additional growth factors associated with tissue repair.
Studies in murine models have also shown that ILC2s can interact with other regulatory cells, such as T reg cells, to suppress inflammation and promote healing. For example, ILC2 responses led to the accumulation of T reg cells and alternatively activated macrophages that protected against cerebral malaria in mice (Besnard et al., 2015). Interactions between T reg cells and ILC2s via maresin 1, a docosahexaenoic acid–derived lipid mediator, were also associated with resolution of murine allergic lung inflammation (Krishnamoorthy et al., 2015). A recent study showed that IL-33–driven ILC2 activation was required for ICOS–ICOSL-mediated interactions with between ILC2s and T reg cells during helminth infection (Molofsky et al., 2015). Together, these data suggest that key interactions between T reg cells and ILC2s lead to resolution of inflammation, but further studies will be needed to assess the molecular mechanisms by which T reg cells and ILC2s colocalize and interact and whether these interactions are conserved in humans.
The capacity of ILC2s to suppress inflammation and support tissue repair is complemented by the part they play in maintaining tissue homeostasis, particularly metabolic homeostasis (Fig. 2). Type 1 inflammation is associated with metabolic disorders and obesity, whereas type 2 cytokines underpin metabolic health, optimized caloric expenditure, and a healthy weight (Wu et al., 2011; Odegaard and Chawla, 2013). Initial work showed that IL-33–responsive, IL-5–producing ILC2s maintained adipose eosinophil and macrophage populations associated with leanness in mice (Molofsky et al., 2013). Subsequent work has highlighted the importance of the IL-33–ILC2 axis specifically in the fat, as ILC2s were enriched in human and murine fat and IL-33–responsive ILC2 responses elicited beige adipocytes that expressed uncoupling protein 1, a key factor in regulating caloric expenditure and limiting adiposity, through expression of met-enkephalin peptides (Brestoff et al., 2015; Lee et al., 2015). Additionally, augmentation of murine ILC2 function by IL-25 and IL-33 treatment could promote weight loss, eosinophil accumulation in the fat, and improved glucose tolerance (Brestoff et al., 2015), and this process was antagonized by proinflammatory IFN-γ responses (Molofsky et al., 2015).
These studies suggest that ILC2s directly and indirectly interact with a variety of cell types in adipose tissue to support metabolic homeostasis. However, the factors that regulate these activities remain incompletely described. Nussbaum et al. (2013) have shown that a neuropeptide regulated by caloric intake, vasoactive intestinal peptide, increased ILC2 production of IL-5, which enhances eosinophil responses associated with the lean state in mice. The ability of ILC2s to sense vitamin A deficiency also suggests that ILC2s can respond to dietary factors to participate in maintaining metabolic homeostasis (Spencer et al., 2014). A very recent study has highlighted the critical importance of cell-intrinsic metabolism in ILC2s (Monticelli et al., 2016). In this study, human and murine ILC2s were shown to express arginase, and cellular bioenergetics controlled by arginine metabolism mediated ILC2 proliferation and production of type 2 cytokines and their ability to promote allergic lung inflammation in mice (Monticelli et al., 2016). Together, these studies bring up fascinating questions regarding how factors in the diet and environment regulate ILC2-mediated metabolic homeostasis and how ILC2-intrinsic metabolism contributes to diverse ILC2 functions. Ultimately, integrated biochemical and immunological approaches will be needed to identify pathways that influence the balance of prohomeostatic and inflammatory ILC2 responses to support concurrent regulation of metabolism, immunity, and inflammation in mice and humans.
The emerging importance of T-bet–expressing ILCs and ILC1s
The characterization of the non-NK cell ILC subset analogous to Th1 cells, the T-bet–expressing ILCs, remained elusive for several years after the identification of ILC2s and ILC3s (Diefenbach et al., 2014; McKenzie et al., 2014; Artis and Spits, 2015). Although the existence of T-bet– and IFN-γ–expressing noncytolytic, helper-like ILC1s and their contribution to type 1 inflammation seemed likely, how ILC1s differed from classical NK cells and other ILC subsets, how these cells developed, and how they contributed to immune responses remained unclear until relatively recently. Here, we will distinguish and discuss the T-bet–expressing ILCs in two groups: T-bet–expressing ILCs that previously expressed RORγt and the T-bet–expressing ILC1s that lack a history of RORγt expression.
T-bet–expressing “ex-ILC3s” and ILC1 development and function
An early study by Vonarbourg et al. (2010) showed that murine NCR−RORγt+ ILCs could lose RORγt expression and acquire the ability to produce IFN-γ in response to IL-12 and IL-15 and that these cells could drive colitis. In the same year, Cella et al. (2010) showed that a subset of human NCR-expressing RORγt+ ILC3s could acquire the ability to produce IFN-γ in response to cytokine signals. Although these studies did not measure expression of T-bet in these IFN-γ–producing ILCs (Cella et al., 2010; Vonarbourg et al., 2010), these data suggest that functional plasticity exists in the ILC3 lineage and that a T-bet– and IFN-γ–expressing ILC1 population can arise from an ILC3 population. Indeed, experiments by Bernink et al. (2013) described ILCs in human tonsil and intestine that expressed T-bet, responded to IL-12 to produce IFN-γ, were enriched in the intestine of patients with Crohn’s disease, and could differentiate from RORγt-expressing ILC3-like cells. In addition, T-bet expression in ILC3s, associated with RORγt down-regulation, can drive acquisition of NKp46 and IFN-γ expression, mediating intestinal inflammation and homeostasis during S. enterica infection in mice (though these particular ILCs appear to be more ILC3-like in vivo; Klose et al., 2013). Together, these data highlight a functionally unique niche for murine and human T-bet–expressing ILCs that previously expressed RORγt, now referred to as “ex-ILC3s” (Fig. 3).
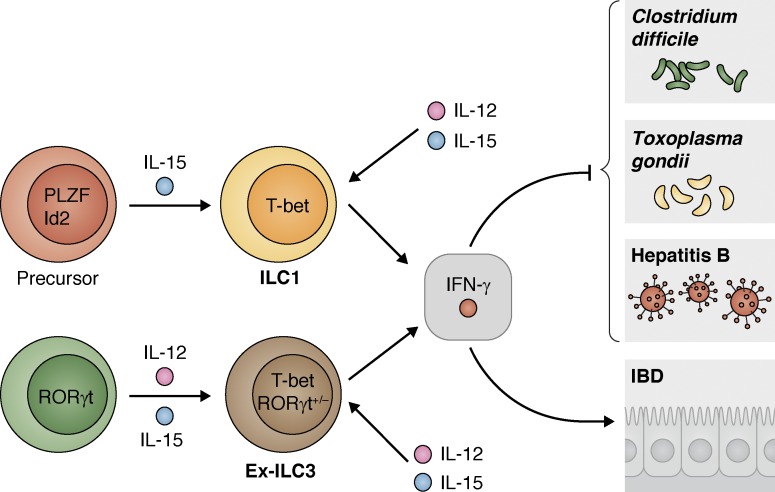
Figure 3.
ILC1s express T-bet and IFN-γ and contribute to type 1 inflammation. T-bet–expressing ILC1s develop from a progenitor that expresses PLZF and Id2 and responds to IL-15. RORγt-expressing cells can also lose expression of RORγt and gain expression of T-bet in response to IL-12 signaling (ex-ILC3s). T-bet–expressing ILCs with a history of RORγt expression or T-bet+ ILC1s respond to IL-12 and IL-15 to produce IFN-γ that is protective during infection with pathogens such as C. difficile, T. gondii, and hepatitis B (not depicted: T-bet–expressing ILCs that currently or previously expressed RORγt are specifically important during infection with S. enterica as well, though these ILCs are more ILC3 like) and may contribute to inflammation and pathology in the intestine in the context of IBD.
More recent work has more clearly defined the development and function of T-bet–expressing ILC1s that likely lack a history of RORγt expression (Fig. 3). Experiments from Fuchs et al. (2013) identified an intraepithelial ILC1 population in humans and mice that responded to IL-12 and IL-15 to produce IFN-γ. These cells were enriched during human and murine colitis and developed independently of RORγt in mice (Fuchs et al., 2013). Constantinides et al. (2014) described a dedicated ILC precursor that expressed PLZF in mice and could differentiate into ILC1s, ILC2s, and ILC3s but not classical NK cells or LTi cells. Similarly, seminal work from Klose et al. (2014) defined an Id2-dependent ILC precursor in mice, the common progenitor to all helper-like ILCs (ChILP), that differentiated into ILC2s and ILC3s and gave rise to T-bet– and IFN-γ–expressing ILC1s that protected against the intracellular protozoan parasite Toxoplasma gondii. A very recent study has shown that the transcription factor Runx3 controls murine ILC1 development and survival (Ebihara et al., 2015). Functionally, murine ILC1s are important in mediating immunity to T. gondii (Klose et al., 2014), human ILC1s are increased in patients with chronic hepatitis B infection (Yang et al., 2015b), and distinct tissue-resident ILC1-like cells are mobilized in response to cell transformation in murine models (Dadi et al., 2016). These data indicate that T-bet–expressing ILC1s that lack a history of RORγt expression are developmentally and functionally distinct ILCs that contribute to immunity to specific pathogens and tumor surveillance in both mice and humans (Fig. 3).
Evolving questions in ILC1 biology
Studies using transcription factor reporter mice, murine models of infection, and human ILCs in vitro and ex vivo have increased our understanding of ILC1 function (Cella et al., 2010; Bernink et al., 2013; Fuchs et al., 2013; Constantinides et al., 2014, 2015; Klose et al., 2014). However, significant questions remain. For example, the molecular nature of the connection between ILC3s and ILC1s that results in the formation of ex-ILC3s in response to environmental cues is not fully defined. Future studies using fate-mapping reporter mice to track the life history of RORγt- and T-bet–expressing ILCs in T– and B cell–sufficient mice will be required to determine how plasticity is regulated and how it affects immune responses. Interestingly, recent work from the Immunological Genome Consortium showed that there was significant overlap in transcriptional profiles of murine ILC1-like cells as compared with classical NK cells and ILC3s (Robinette et al., 2015), and expression of eomesodermin distinguishes ILC1s and classical NK cells during development (Gordon et al., 2012; Daussy et al., 2014; Klose et al., 2014; Pikovskaya et al., 2016). A better understanding of the relationship between ILC1s and ILC3s will be key in deciphering the significance of T-bet–expressing cells that previously expressed RORγt in various contexts.
Although it has been assumed that ILC1s functionally resemble Th1 cells, their contribution to various inflammatory states has not been fully explored. Based on their association with Crohn’s disease in humans (Bernink et al., 2013; Fuchs et al., 2013), it seems likely that ILC1s are involved in the pathogenesis of IBD. In addition, as IBD is strongly associated with dysregulation of the relationship between the host and the microbiota (Alexander et al., 2014), ILC1s can likely integrate cues from the microbiota or damaged epithelial cells to modulate inflammatory responses, though whether murine or human ILC1s have this ability and its significance remain unclear. In addition to their potential role in intestinal inflammation, ILC1s likely contribute to inflammation at other tissue sites. ILC1s that express CD4 are increased in humans with systemic sclerosis (Roan et al., 2016), and circulating IL-22 and ATP levels are increased in patients after liver resection, with ILC1s being a potent source of IL-22 in this context (Kudira et al., 2016). These data suggest that ILC1s play a role in various autoimmune and inflammatory conditions. Notably, a recent study showed that murine NKp46+ ILC1s in the vaginal mucosa actually dampened CD8+ T cell responses after local vaccination, suggesting that ILC1s could also have antiinflammatory properties (Luci et al., 2015).
In addition, ILC1s likely contribute to immune responses to various viral, bacterial, and protozoan pathogens. Although we know that transferred T-bet–expressing ILC1s that developed from the ChILP provided protection from T. gondii infection in Rag2−/−Il2rγ−/− mice (Klose et al., 2014), how these cells contribute to protective inflammation and immune-mediated pathology after infection with diverse pathogens, including other protozoans and intracellular bacterial pathogens, is unclear. A recent key study has highlighted the critical importance of T-bet–expressing ILC1s, with ILC3s playing a minor role, in antibacterial immune responses after infection with Clostridium difficile (Abt et al., 2015), but much remains to be discovered regarding the regulation and function of these cells and T-bet–expressing ILCs with a history of RORγt expression during bacterial and, particularly, viral infection. Collectively, the studies of T-bet–expressing ILCs to date have begun to elucidate their development, lineage relationship to other ILCs, and functional capacities, but much still needs to be done to create a comprehensive phenotypic and functional profile for murine and human ILC1s.
The next challenges: Targeting ILCs in human diseases
The critical roles that ILCs play in a myriad of disease states suggest that novel therapeutics that target these cells could improve outcomes for inflammatory diseases in humans (Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015; Goldberg et al., 2015; Peebles, 2015). The emphasis that recent studies have placed on the biology of human ILCs reflects interest in exploring this possibility. Although there are multiple disease states that could be treated by modulating ILC function, here we briefly highlight potential opportunities to therapeutically target ILC activities in two human diseases, IBD and asthma, and discuss the challenges that may be encountered in the course of developing drugs in this context.
The close association of ILC3s with intestinal inflammation has prompted long-standing interest in the role of ILC3s in IBD (McKenzie et al., 2014; Artis and Spits, 2015; Goldberg et al., 2015). Importantly, ILC3s are enriched or their phenotypes are altered in the inflamed intestine of patients with IBD (Geremia et al., 2011; Bernink et al., 2013; Fuchs et al., 2013; Hepworth et al., 2015). Together with data from murine models, these studies support the idea that ILC3s might be targeted to treat IBD (Goldberg et al., 2015). However, two major challenges exist when considering pursuing this approach. First, ILC3s are rich sources of IL-22, which can be protective or pathogenic in the intestine (Zenewicz et al., 2008; Sonnenberg et al., 2011a; Huber et al., 2012; Kirchberger et al., 2013). Thus, whether changes in IL-22–producing ILC3 populations in the intestine of IBD patients are a cause or effect of intestinal inflammation remains unclear. Second, the importance of the relationships between intestinal ILC3s, the microbiota, and other immune cells (Eberl, 2012; Tait Wojno and Artis, 2012; McKenzie et al., 2014; Artis and Spits, 2015) suggests that modulating ILC3 function could have off-target effects on the host–microbiota relationship, though evidence suggests that NCR-expressing ILC3 deficiency does not affect the composition of the microbiota in mice (Rankin et al., 2016). Notably, a recent study in mice has shown that transient inhibition of Rorγt in murine models of intestinal inflammation spares beneficial ILC3 function and specifically targets pathogenic Th17 responses (Withers et al., 2016). These findings suggest that certain pathways associated with ILC3 function can be targeted without compromising the tissue-protective functions of these cells (Withers et al., 2016) or that differences in the spacial, temporal, or functional distribution of T cells and NCR+ and NCR− ILC3 subsets could be leveraged to develop specific therapies. Additional research in preclinical models and examining human intestinal ILC3s in healthy individuals and through a time course of disease and after treatment in IBD patients will be needed to determine whether ILC3s and their regulatory pathways are appropriate targets to treat IBD in humans.
The importance of ILC2s in allergic airway disease was first highlighted in murine models (Halim et al., 2012a; Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015), and recent studies have revealed an association between ILC2s and asthma in humans (Bartemes et al., 2014; Castanhinha et al., 2015; Christianson et al., 2015). Thus, ILC2s have been proposed as targets for therapeutic intervention in various asthma endotypes (Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015; Peebles, 2015). Notably, recent work has shown that pro-allergic ILC2 responses are regulated by eicosanoids (Barnig et al., 2013; Doherty et al., 2013; Kim et al., 2013b; McKenzie et al., 2014; Artis and Spits, 2015; Peebles, 2015; Tait Wojno et al., 2015), suggesting that ILC2 function could be modulated indirectly by manipulating eicosanoid pathways (Pettipher and Whittaker, 2012). Specifically, as the PGD2–CRTH2 pathway is associated with ILC2 activation (Barnig et al., 2013; Xue et al., 2014; Tait Wojno et al., 2015), inhibition of this axis could be effective, and indeed, some clinical trials have shown that CRTH2 inhibition can decrease asthma symptoms, though this is not uniformly effective (Pettipher and Whittaker, 2012). In addition, as LXA4 can limit human ILC2 responses and is decreased in patients with severe asthma, treatment with exogenous LXA4 or agonism of its receptor may also be a possibility (Peebles, 2015). In addition, recent work showing that arginase is expressed by human and murine ILC2s and mediates their function in allergic pulmonary inflammation suggests that the ILC2–arginase axis could be modulated to treat allergic asthma in humans (Monticelli et al., 2016), particularly in light of previous work validating the arginine pathway as a potential therapeutic target in human disease (Scott and Grasemann, 2014). Thus, efforts to regulate ILC2 activities to treat asthma are already underway in some respects. However, significant challenges remain. In particular, targeting ILC2s in the lung may have unwanted side effects, as these cells also mediate tissue repair (Monticelli et al., 2011; Turner et al., 2013). Second, inhibiting factors upstream of ILC2 responses may impact other cell types as well, and further work will be needed to determine whether targeting ILC2s or the factors that regulate them will be effective and safe, though recent studies showing that type 1 IFN and IL-27 limit pathogenic ILC2 functions (Duerr et al., 2016; Mchedlidze et al., 2016; Moro et al., 2016) suggest that we have much to learn about ILC2 regulation and that proinflammatory ILC2 activities could be limited by enhancing pathways that shut down these cells. Finally, although important studies have been performed to characterize ILC2 biology in the human lung (Mjösberg et al., 2011; Monticelli et al., 2011, 2016; Hams et al., 2014; Tait Wojno et al., 2015; De Grove et al., 2016), how mechanistic data derived from studies of murine models of allergic respiratory disease will translate into studies of human patient populations remains unknown. Thus, studies in preclinical models that closely reflect human asthma phenotypes and endotypes and extensive characterization of ILC responses in diverse groups of asthma patients will be required to validate ILC2s as a target in asthma treatment.
The examples of IBD and asthma provide potential opportunities to modulate ILC function to treat inflammatory diseases that cause significant morbidity. The challenges that exist in this endeavor are formidable, and a common theme is the difficulty inherent in selectively targeting ILCs (McKenzie et al., 2014; Artis and Spits, 2015; Goldberg et al., 2015; Peebles, 2015). However, a recent study in which ILC3 populations were unexpectedly selectively depleted in multiple sclerosis patients who were treated with an anti-CD25 antibody (Perry et al., 2012) suggests that there are pathways that can be exploited to modulate ILCs. However, preclinical models will again be required to validate whether certain pathways that influence, but are not specific to, ILCs might be appropriate for therapeutic targeting. Preclinical models used in conjunction with genome-wide profiling may also be useful in identifying truly unique pathways that could be modulated to alter ILC responses. In addition, detailed immunological analyses of patient responses after treatment with an array of monoclonal antibodies and small molecules will be critical in identifying whether any preexisting therapies selectively regulate ILCs. Ultimately, the complex nature of the relationship between ILCs, the epithelium, the microbiota, and other immune cells suggests that altering ILC function to treat inflammatory disease will be challenging. Conversely, the placement of ILCs at the center of such a powerful immunological communication network also makes these cells a unique target in the treatment of inflammatory diseases.
Acknowledgments
The authors wish to thank members of the Tait Wojno laboratory, the Artis laboratory, the Sonnenberg laboratory, and Chalya Pudlewski for discussions and critical reading of the manuscript.
Research in the Tait Wojno laboratory is supported by the National Institutes of Health (K22-AI116729) and Cornell University. Research in the Artis laboratory is supported by the National Institutes of Health (AI061570, AI095608, AI087990, AI074878, AI095466, AI106697, AI102942, and AI097333 to D. Artis), the Crohn’s and Colitis Foundation of America (D. Artis), the Burroughs Wellcome Fund Investigator in Pathogenesis of Infectious Disease Award (D. Artis), and the National Institute of Allergy and Infectious Diseases Mucosal Immunology Studies Team (MIST) consortium (U01-AI095608).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AhR
- aryl hydrocarbon receptor
- Areg
- amphiregulin
- GALT
- gut-associated lymphoid tissue
- IBD
- inflammatory bowel disease
- ICOS
- inducible T cell costimulator
- ICOSL
- ICOS ligand
- ILC
- innate lymphoid cell
- ILF
- isolated lymphoid follicle
- LTi
- lymphoid tissue inducer
- PGD2
- prostaglandin D2
- PLZF
- promyelocytic leukemia zinc finger protein
- ROR
- retinoid-related orphan receptor
- TSLP
- thymic stromal lymphopoietin
References
- Abt M.C., Lewis B.B., Caballero S., Xiong H., Carter R.A., Sušac B., Ling L., Leiner I., and Pamer E.G.. 2015. Innate immune defenses Mediated by two ILC subsets are critical for protection against acute Clostridium difficile infection. Cell Host Microbe. 18:27–37. 10.1016/j.chom.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abt M.C., Buffie C.G., Sušac B., Becattini S., Carter R.A., Leiner I., Keith J.W., Artis D., Osborne L.C., and Pamer E.G.. 2016. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci. Transl. Med. 8:327ra25 10.1126/scitranslmed.aad6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander K.L., Targan S.R., and Elson C.O. III. 2014. Microbiota activation and regulation of innate and adaptive immunity. Immunol. Rev. 260:206–220. 10.1111/imr.12180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D., and Spits H.. 2015. The biology of innate lymphoid cells. Nature. 517:293–301. 10.1038/nature14189 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Ando M., Kamada N., Nagano Y., Narushima S., Suda W., Imaoka A., Setoyama H., Nagamori T., et al. . 2015. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 163:367–380. 10.1016/j.cell.2015.08.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla S.J., Chan Y.R., Zheng M., Fei M., Askew D.J., Pociask D.A., Reinhart T.A., McAllister F., Edeal J., Gaus K., et al. . 2008. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14:275–281. 10.1038/nm1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerenwaldt A., von Burg N., Kreuzaler M., Sitte S., Horvath E., Peter A., Voehringer D., Rolink A.G., and Finke D.. 2016. Flt3 ligand regulates the development of innate lymphoid cells in fetal and adult mice. J. Immunol. 196:2561–2571. 10.4049/jimmunol.1501380 [DOI] [PubMed] [Google Scholar]
- Bando J.K., Liang H.E., and Locksley R.M.. 2015. Identification and distribution of developing innate lymphoid cells in the fetal mouse intestine. Nat. Immunol. 16:153–160. 10.1038/ni.3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnig C., Cernadas M., Dutile S., Liu X., Perrella M.A., Kazani S., Wechsler M.E., Israel E., and Levy B.D.. 2013. Lipoxin A4 regulates natural killer cell and type 2 innate lymphoid cell activation in asthma. Sci. Transl. Med. 5:174ra26 10.1126/scitranslmed.3004812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartemes K.R., Kephart G.M., Fox S.J., and Kita H.. 2014. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J. Allergy Clin. Immunol. 134:671–678.e4. 10.1016/j.jaci.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink J.H., Peters C.P., Munneke M., te Velde A.A., Meijer S.L., Weijer K., Hreggvidsdottir H.S., Heinsbroek S.E., Legrand N., Buskens C.J., et al. . 2013. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat. Immunol. 14:221–229. 10.1038/ni.2534 [DOI] [PubMed] [Google Scholar]
- Besnard A.G., Guabiraba R., Niedbala W., Palomo J., Reverchon F., Shaw T.N., Couper K.N., Ryffel B., and Liew F.Y.. 2015. IL-33-mediated protection against experimental cerebral malaria is linked to induction of type 2 innate lymphoid cells, M2 macrophages and regulatory T cells. PLoS Pathog. 11:e1004607 10.1371/journal.ppat.1004607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund A.K., Forkel M., Picelli S., Konya V., Theorell J., Friberg D., Sandberg R., and Mjösberg J.. 2016. The heterogeneity of human CD127+ innate lymphoid cells revealed by single-cell RNA sequencing. Nat. Immunol. 17:451–460. 10.1038/ni.3368 [DOI] [PubMed] [Google Scholar]
- Bouchery T., Kyle R., Camberis M., Shepherd A., Filbey K., Smith A., Harvie M., Painter G., Johnston K., Ferguson P., et al. . 2015. ILC2s and T cells cooperate to ensure maintenance of M2 macrophages for lung immunity against hookworms. Nat. Commun. 6:6970 10.1038/ncomms7970 [DOI] [PubMed] [Google Scholar]
- Bouskra D., Brézillon C., Bérard M., Werts C., Varona R., Boneca I.G., and Eberl G.. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 456:507–510. 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- Brestoff J.R., Kim B.S., Saenz S.A., Stine R.R., Monticelli L.A., Sonnenberg G.F., Thome J.J., Farber D.L., Lutfy K., Seale P., and Artis D.. 2015. Group 2 innate lymphoid cells promote beiging of white adipose tissue and limit obesity. Nature. 519:242–246. 10.1038/nature14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonocore S., Ahern P.P., Uhlig H.H., Ivanov I.I., Littman D.R., Maloy K.J., and Powrie F.. 2010. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 464:1371–1375. 10.1038/nature08949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Califano D., Cho J.J., Uddin M.N., Lorentsen K.J., Yang Q., Bhandoola A., Li H., and Avram D.. 2015. Transcription factor Bcl11b controls identity and function of mature type 2 innate lymphoid cells. Immunity. 43:354–368. 10.1016/j.immuni.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrega P., Loiacono F., Di Carlo E., Scaramuccia A., Mora M., Conte R., Benelli R., Spaggiari G.M., Cantoni C., Campana S., et al. . 2015. NCR+ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat. Commun. 6:8280 10.1038/ncomms9280 [DOI] [PubMed] [Google Scholar]
- Castanhinha S., Sherburn R., Walker S., Gupta A., Bossley C.J., Buckley J., Ullmann N., Grychtol R., Campbell G., Maglione M., et al. . 2015. Pediatric severe asthma with fungal sensitization is mediated by steroid-resistant IL-33. J. Allergy Clin. Immunol. 136:312–322.e7. 10.1016/j.jaci.2015.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella M., Otero K., and Colonna M.. 2010. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1β reveals intrinsic functional plasticity. Proc. Natl. Acad. Sci. USA. 107:10961–10966. 10.1073/pnas.1005641107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan I.H., Jain R., Tessmer M.S., Gorman D., Mangadu R., Sathe M., Vives F., Moon C., Penaflor E., Turner S., et al. . 2014. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol. 7:842–856. 10.1038/mi.2013.101 [DOI] [PubMed] [Google Scholar]
- Chen J., Waddell A., Lin Y.D., and Cantorna M.T.. 2015a Dysbiosis caused by vitamin D receptor deficiency confers colonization resistance to Citrobacter rodentium through modulation of innate lymphoid cells. Mucosal Immunol. 8:618–626. 10.1038/mi.2014.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., He Z., Slinger E., Bongers G., Lapenda T.L., Pacer M.E., Jiao J., Beltrao M.F., Soto A.J., Harpaz N., et al. . 2015b IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol. 8:390–402. 10.1038/mi.2014.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier M., Sawa S., and Eberl G.. 2012. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209:729–740. 10.1084/jem.20111594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson C.A., Goplen N.P., Zafar I., Irvin C., Good J.T. Jr., Rollins D.R., Gorentla B., Liu W., Gorska M.M., Chu H., et al. . 2015. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J. Allergy Clin. Immunol. 136:59–68.e14. 10.1016/j.jaci.2014.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cols M., Rahman A., Maglione P.J., Garcia-Carmona Y., Simchoni N., Ko H.B., Radigan L., Cerutti A., Blankenship D., Pascual V., and Cunningham-Rundles C.. 2016. Expansion of inflammatory innate lymphoid cells in patients with common variable immune deficiency. J. Allergy Clin. Immunol. 137:1206–1215.e6. 10.1016/j.jaci.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401. 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides M.G., Gudjonson H., McDonald B.D., Ishizuka I.E., Verhoef P.A., Dinner A.R., and Bendelac A.. 2015. PLZF expression maps the early stages of ILC1 lineage development. Proc. Natl. Acad. Sci. USA. 112:5123–5128. 10.1073/pnas.1423244112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T., Lund F.E., Ngo V.N., Randall T.D., Jansen W., Greuter M.J., de Waal-Malefyt R., Kraal G., Cyster J.G., and Mebius R.E.. 2004. Initiation of cellular organization in lymph nodes is regulated by non-B cell-derived signals and is not dependent on CXC chemokine ligand 13. J. Immunol. 173:4889–4896. 10.4049/jimmunol.173.8.4889 [DOI] [PubMed] [Google Scholar]
- Cupedo T., Crellin N.K., Papazian N., Rombouts E.J., Weijer K., Grogan J.L., Fibbe W.E., Cornelissen J.J., and Spits H.. 2009. Human fetal lymphoid tissue–inducer cells are interleukin 17–producing precursors to RORC+ CD127+ natural killer–like cells. Nat. Immunol. 10:66–74. 10.1038/ni.1668 [DOI] [PubMed] [Google Scholar]
- Dadi S., Chhangawala S., Whitlock B.M., Franklin R.A., Luo C.T., Oh S.A., Toure A., Pritykin Y., Huse M., Leslie C.S., and Li M.O.. 2016. Cancer immunosurveillance by tissue-resident innate lymphoid cells and innate-like T cells. Cell. 164:365–377. 10.1016/j.cell.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy C., Faure F., Mayol K., Viel S., Gasteiger G., Charrier E., Bienvenu J., Henry T., Debien E., Hasan U.A., et al. . 2014. T-bet and Eomes instruct the development of two distinct natural killer cell lineages in the liver and in the bone marrow. J. Exp. Med. 211:563–577. 10.1084/jem.20131560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grove K.C., Provoost S., Verhamme F.M., Bracke K.R., Joos G.F., Maes T., and Brusselle G.G.. 2016. Characterization and quantification of innate lymphoid cell subsets in human lung. PLoS One. 11:e0145961 10.1371/journal.pone.0145961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denney L., Byrne A.J., Shea T.J., Buckley J.S., Pease J.E., Herledan G.M., Walker S.A., Gregory L.G., and Lloyd C.M.. 2015. Pulmonary epithelial cell-derived cytokine TGF-β1 is a critical cofactor for enhanced innate lymphoid cell function. Immunity. 43:945–958. 10.1016/j.immuni.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diefenbach A., Colonna M., and Koyasu S.. 2014. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 41:354–365. 10.1016/j.immuni.2014.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty T.A., Khorram N., Lund S., Mehta A.K., Croft M., and Broide D.H.. 2013. Lung type 2 innate lymphoid cells express cysteinyl leukotriene receptor 1, which regulates TH2 cytokine production. J. Allergy Clin. Immunol. 132:205–213. 10.1016/j.jaci.2013.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty T.A., Baum R., Newbury R.O., Yang T., Dohil R., Aquino M., Doshi A., Walford H.H., Kurten R.C., Broide D.H., and Aceves S.. 2015. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J. Allergy Clin. Immunol. 136:792–794.e3. 10.1016/j.jaci.2015.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudakov J.A., Hanash A.M., Jenq R.R., Young L.F., Ghosh A., Singer N.V., West M.L., Smith O.M., Holland A.M., Tsai J.J., et al. . 2012. Interleukin-22 drives endogenous thymic regeneration in mice. Science. 336:91–95. 10.1126/science.1218004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerr C.U., McCarthy C.D., Mindt B.C., Rubio M., Meli A.P., Pothlichet J., Eva M.M., Gauchat J.F., Qureshi S.T., Mazer B.D., et al. . 2016. Type I interferon restricts type 2 immunopathology through the regulation of group 2 innate lymphoid cells. Nat. Immunol. 17:65–75. 10.1038/ni.3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G. 2012. Development and evolution of RORγt+ cells in a microbe’s world. Immunol. Rev. 245:177–188. 10.1111/j.1600-065X.2011.01071.x [DOI] [PubMed] [Google Scholar]
- Eberl G., and Littman D.R.. 2004. Thymic origin of intestinal αβ T cells revealed by fate mapping of RORγt+ cells. Science. 305:248–251. 10.1126/science.1096472 [DOI] [PubMed] [Google Scholar]
- Eberl G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73. 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Ebihara T., Song C., Ryu S.H., Plougastel-Douglas B., Yang L., Levanon D., Groner Y., Bern M.D., Stappenbeck T.S., Colonna M., et al. . 2015. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 16:1124–1133. 10.1038/ni.3272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenring M., vom Berg J., Kristiansen G., Saller E., and Becher B.. 2010. IL-12 initiates tumor rejection via lymphoid tissue-inducer cells bearing the natural cytotoxicity receptor NKp46. Nat. Immunol. 11:1030–1038. 10.1038/ni.1947 [DOI] [PubMed] [Google Scholar]
- Fallon P.G., Ballantyne S.J., Mangan N.E., Barlow J.L., Dasvarma A., Hewett D.R., McIlgorm A., Jolin H.E., and McKenzie A.N.. 2006. Identification of an interleukin (IL)-25–dependent cell population that provides IL-4, IL-5, and IL-13 at the onset of helminth expulsion. J. Exp. Med. 203:1105–1116. 10.1084/jem.20051615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A., Vermi W., Lee J.S., Lonardi S., Gilfillan S., Newberry R.D., Cella M., and Colonna M.. 2013. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 38:769–781. 10.1016/j.immuni.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.C., Bessman N.J., Hepworth M.R., Kumar N., Shibata N., Kobuley D., Wang K., Ziegler C.G., Goc J., Shima T., et al. . 2016. Lymphoid-tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity. 44:634–646. 10.1016/j.immuni.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant M.A., Slipetz D., Hamelin E., Rochdi M.D., Talbot S., de Brum-Fernandes A.J., and Parent J.L.. 2007. Differential regulation of the signaling and trafficking of the two prostaglandin D2 receptors, prostanoid DP receptor and CRTH2. Eur. J. Pharmacol. 557:115–123. 10.1016/j.ejphar.2006.11.058 [DOI] [PubMed] [Google Scholar]
- Geiger T.L., Abt M.C., Gasteiger G., Firth M.A., O’Connor M.H., Geary C.D., O’Sullivan T.E., van den Brink M.R., Pamer E.G., Hanash A.M., and Sun J.C.. 2014. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J. Exp. Med. 211:1723–1731. 10.1084/jem.20140212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F., Sidot E., Smyth D.J., Ohmoto M., Matsumoto I., Dardalhon V., Cesses P., Garnier L., Pouzolles M., Brulin B., et al. . 2016. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 529:226–230. 10.1038/nature16527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia A., Arancibia-Cárcamo C.V., Fleming M.P., Rust N., Singh B., Mortensen N.J., Travis S.P., and Powrie F.. 2011. IL-23–responsive innate lymphoid cells are increased in inflammatory bowel disease. J. Exp. Med. 208:1127–1133. 10.1084/jem.20101712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomin P.R., Moy R.H., Noti M., Osborne L.C., Siracusa M.C., Alenghat T., Liu B., McCorkell K.A., Troy A.E., Rak G.D., et al. . 2015. Epithelial-intrinsic IKKα expression regulates group 3 innate lymphoid cell responses and antibacterial immunity. J. Exp. Med. 212:1513–1528. 10.1084/jem.20141831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladiator A., Wangler N., Trautwein-Weidner K., and LeibundGut-Landmann S.. 2013. Cutting edge: IL-17-secreting innate lymphoid cells are essential for host defense against fungal infection. J. Immunol. 190:521–525. 10.4049/jimmunol.1202924 [DOI] [PubMed] [Google Scholar]
- Goldberg R., Prescott N., Lord G.M., MacDonald T.T., and Powell N.. 2015. The unusual suspects—innate lymphoid cells as novel therapeutic targets in IBD. Nat. Rev. Gastroenterol. Hepatol. 12:271–283. 10.1038/nrgastro.2015.52 [DOI] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., and Reiner S.L.. 2012. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 36:55–67. 10.1016/j.immuni.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y., Obata T., Kunisawa J., Sato S., Ivanov I.I., Lamichhane A., Takeyama N., Kamioka M., Sakamoto M., Matsuki T., et al. . 2014. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science. 345:1254009 10.1126/science.1254009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., Krauss R.H., Sun A.C., and Takei F.. 2012a Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity. 36:451–463. 10.1016/j.immuni.2011.12.020 [DOI] [PubMed] [Google Scholar]
- Halim T.Y., MacLaren A., Romanish M.T., Gold M.J., McNagny K.M., and Takei F.. 2012b Retinoic-acid-receptor-related orphan nuclear receptor alpha is required for natural helper cell development and allergic inflammation. Immunity. 37:463–474. 10.1016/j.immuni.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Halim T.Y., Steer C.A., Mathä L., Gold M.J., Martinez-Gonzalez I., McNagny K.M., McKenzie A.N., and Takei F.. 2014. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 40:425–435. 10.1016/j.immuni.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hams E., Armstrong M.E., Barlow J.L., Saunders S.P., Schwartz C., Cooke G., Fahy R.J., Crotty T.B., Hirani N., Flynn R.J., et al. . 2014. IL-25 and type 2 innate lymphoid cells induce pulmonary fibrosis. Proc. Natl. Acad. Sci. USA. 111:367–372. 10.1073/pnas.1315854111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth M.R., Monticelli L.A., Fung T.C., Ziegler C.G., Grunberg S., Sinha R., Mantegazza A.R., Ma H.L., Crawford A., Angelosanto J.M., et al. . 2013. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 498:113–117. 10.1038/nature12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth M.R., Fung T.C., Masur S.H., Kelsen J.R., McConnell F.M., Dubrot J., Withers D.R., Hugues S., Farrar M.A., Reith W., et al. . 2015. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science. 348:1031–1035. 10.1126/science.aaa4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández P.P., Mahlakõiv T., Yang I., Schwierzeck V., Nguyen N., Guendel F., Gronke K., Ryffel B., Hölscher C., Dumoutier L., et al. . 2015. Interferon-λ and interleukin 22 act synergistically for the induction of interferon-stimulated genes and control of rotavirus infection. Nat. Immunol. 16:698–707. 10.1038/ni.3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitt M.R., Lavoie S., Michaud M., Blum A.M., Tran S.V., Weinstock J.V., Gallini C.A., Redding K., Margolskee R.F., Osborne L.C., et al. . 2016. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 351:1329–1333. 10.1126/science.aaf1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler T., Klose C.S., Souabni A., Turqueti-Neves A., Pfeifer D., Rawlins E.L., Voehringer D., Busslinger M., and Diefenbach A.. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 37:634–648. 10.1016/j.immuni.2012.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Niu Z., Tan J., Yang J., Liu Y., Ma H., Lee V.W., Sun S., Song X., Guo M., et al. . 2015a IL-25 elicits innate lymphoid cells and multipotent progenitor type 2 cells that reduce renal ischemic/reperfusion injury. J. Am. Soc. Nephrol. 26:2199–2211. 10.1681/ASN.2014050479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Guo L., Qiu J., Chen X., Hu-Li J., Siebenlist U., Williamson P.R., Urban J.F. Jr., and Paul W.E.. 2015b IL-25-responsive, lineage-negative KLRG1hi cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat. Immunol. 16:161–169. 10.1038/ni.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Zenewicz L.A., Huber F.J., Bosurgi L., Hu B., Hedl M., Zhang W., O’Connor W. Jr., Murphy A.J., et al. . 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 491:259–263. 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka I.E., Chea S., Gudjonson H., Constantinides M.G., Dinner A.R., Bendelac A., and Golub R.. 2016. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat. Immunol. 17:269–276. 10.1038/ni.3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi F., Isshiki T., Harada N., Akiba H., and Miyake S.. 2015. ICOS promotes group 2 innate lymphoid cell activation in lungs. Biochem. Biophys. Res. Commun. 463:739–745. 10.1016/j.bbrc.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Kanamori Y., Ishimaru K., Nanno M., Maki K., Ikuta K., Nariuchi H., and Ishikawa H.. 1996. Identification of novel lymphoid tissues in murine intestinal mucosa where clusters of c-kit+ IL-7R+ Thy1+ lympho-hemopoietic progenitors develop. J. Exp. Med. 184:1449–1459. 10.1084/jem.184.4.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearley J., Silver J.S., Sanden C., Liu Z., Berlin A.A., White N., Mori M., Pham T.H., Ward C.K., Criner G.J., et al. . 2015. Cigarette smoke silences innate lymphoid cell function and facilitates an exacerbated type I interleukin-33-dependent response to infection. Immunity. 42:566–579. 10.1016/j.immuni.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Kim B.S., Siracusa M.C., Saenz S.A., Noti M., Monticelli L.A., Sonnenberg G.F., Hepworth M.R., Van Voorhees A.S., Comeau M.R., and Artis D.. 2013a TSLP elicits IL-33-independent innate lymphoid cell responses to promote skin inflammation. Sci. Transl. Med. 5:170ra16 10.1126/scitranslmed.3005374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.S., Wojno E.D., and Artis D.. 2013b Innate lymphoid cells and allergic inflammation. Curr. Opin. Immunol. 25:738–744. 10.1016/j.coi.2013.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.S., Wang K., Siracusa M.C., Saenz S.A., Brestoff J.R., Monticelli L.A., Noti M., Tait Wojno E.D., Fung T.C., Kubo M., and Artis D.. 2014. Basophils promote innate lymphoid cell responses in inflamed skin. J. Immunol. 193:3717–3725. 10.4049/jimmunol.1401307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.H., Taparowsky E.J., and Kim C.H.. 2015. Retinoic acid differentially regulates the migration of innate lymphoid cell subsets to the gut. Immunity. 43:107–119. 10.1016/j.immuni.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S., Royston D.J., Boulard O., Thornton E., Franchini F., Szabady R.L., Harrison O., and Powrie F.. 2013. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 210:917–931. 10.1084/jem.20122308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss E.A., Vonarbourg C., Kopfmann S., Hobeika E., Finke D., Esser C., and Diefenbach A.. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 334:1561–1565. 10.1126/science.1214914 [DOI] [PubMed] [Google Scholar]
- Klose C.S., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., d’Hargues Y., Göppert N., Croxford A.L., Waisman A., Tanriver Y., and Diefenbach A.. 2013. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature. 494:261–265. 10.1038/nature11813 [DOI] [PubMed] [Google Scholar]
- Klose C.S., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. . 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 157:340–356. 10.1016/j.cell.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy N., Burkett P.R., Dalli J., Abdulnour R.E., Colas R., Ramon S., Phipps R.P., Petasis N.A., Kuchroo V.K., Serhan C.N., and Levy B.D.. 2015. Cutting edge: maresin-1 engages regulatory T cells to limit type 2 innate lymphoid cell activation and promote resolution of lung inflammation. J. Immunol. 194:863–867. 10.4049/jimmunol.1402534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglov A.A., Grivennikov S.I., Kuprash D.V., Winsauer C., Prepens S., Seleznik G.M., Eberl G., Littman D.R., Heikenwalder M., Tumanov A.V., and Nedospasov S.A.. 2013. Nonredundant function of soluble LTα3 produced by innate lymphoid cells in intestinal homeostasis. Science. 342:1243–1246. 10.1126/science.1243364 [DOI] [PubMed] [Google Scholar]
- Kudira R., Malinka T., Kohler A., Dosch M., de Agüero M.G., Melin N., Haegele S., Starlinger P., Maharjan N., Saxena S., et al. . 2016. P2X1-regulated IL-22 secretion by innate lymphoid cells is required for efficient liver regeneration. Hepatology. 63:2004–2017. 10.1002/hep.28492 [DOI] [PubMed] [Google Scholar]
- Lee J.S., Cella M., McDonald K.G., Garlanda C., Kennedy G.D., Nukaya M., Mantovani A., Kopan R., Bradfield C.A., Newberry R.D., and Colonna M.. 2012. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13:144–151. 10.1038/ni.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.W., Odegaard J.I., Mukundan L., Qiu Y., Molofsky A.B., Nussbaum J.C., Yun K., Locksley R.M., and Chawla A.. 2015. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 160:74–87. 10.1016/j.cell.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Razumilava N., Gores G.J., Walters S., Mizuochi T., Mourya R., Bessho K., Wang Y.H., Glaser S.S., Shivakumar P., and Bezerra J.A.. 2014. Biliary repair and carcinogenesis are mediated by IL-33-dependent cholangiocyte proliferation. J. Clin. Invest. 124:3241–3251. 10.1172/JCI73742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim A.I., Menegatti S., Bustamante J., Le Bourhis L., Allez M., Rogge L., Casanova J.L., Yssel H., and Di Santo J.P.. 2016. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J. Exp. Med. 213:569–583. 10.1084/jem.20151750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner M., Ohnmacht C., Presley L., Bruhns P., Si-Tahar M., Sawa S., and Eberl G.. 2011. Microbiota-induced tertiary lymphoid tissues aggravate inflammatory disease in the absence of RORγt and LTi cells. J. Exp. Med. 208:125–134. 10.1084/jem.20100052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longman R.S., Diehl G.E., Victorio D.A., Huh J.R., Galan C., Miraldi E.R., Swaminath A., Bonneau R., Scherl E.J., and Littman D.R.. 2014. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J. Exp. Med. 211:1571–1583. 10.1084/jem.20140678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C.A., Kingsbury D.D., Velazquez E.M., and Bäumler A.J.. 2014. Collateral damage: microbiota-derived metabolites and immune function in the antibiotic era. Cell Host Microbe. 16:156–163. 10.1016/j.chom.2014.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci C., Bekri S., Bihl F., Pini J., Bourdely P., Nouhen K., Malgogne A., Walzer T., Braud V.M., and Anjuère F.. 2015. NKp46+ innate lymphoid cells dampen vaginal CD8 T cell responses following local immunization with a cholera toxin-based vaccine. PLoS One. 10:e0143224 10.1371/journal.pone.0143224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maazi H., Patel N., Sankaranarayanan I., Suzuki Y., Rigas D., Soroosh P., Freeman G.J., Sharpe A.H., and Akbari O.. 2015. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity. 42:538–551. 10.1016/j.immuni.2015.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchedlidze T., Waldner M., Zopf S., Walker J., Rankin A.L., Schuchmann M., Voehringer D., McKenzie A.N., Neurath M.F., Pflanz S., and Wirtz S.. 2013. Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity. 39:357–371. 10.1016/j.immuni.2013.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mchedlidze T., Kindermann M., Neves A.T., Voehringer D., Neurath M.F., and Wirtz S.. 2016. IL-27 suppresses type 2 immune responses in vivo via direct effects on group 2 innate lymphoid cells. Mucosal Immunol. 10.1038/mi.2016.20 [DOI] [PubMed] [Google Scholar]
- McKenzie A.N., Spits H., and Eberl G.. 2014. Innate lymphoid cells in inflammation and immunity. Immunity. 41:366–374. 10.1016/j.immuni.2014.09.006 [DOI] [PubMed] [Google Scholar]
- McSorley H.J., Blair N.F., Smith K.A., McKenzie A.N., and Maizels R.M.. 2014. Blockade of IL-33 release and suppression of type 2 innate lymphoid cell responses by helminth secreted products in airway allergy. Mucosal Immunol. 7:1068–1078. 10.1038/mi.2013.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mebius R.E. 2003. Organogenesis of lymphoid tissues. Nat. Rev. Immunol. 3:292–303. 10.1038/nri1054 [DOI] [PubMed] [Google Scholar]
- Mebius R.E., Rennert P., and Weissman I.L.. 1997. Developing lymph nodes collect CD4+CD3− LTβ+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 7:493–504. 10.1016/S1074-7613(00)80371-4 [DOI] [PubMed] [Google Scholar]
- Meylan F., Hawley E.T., Barron L., Barlow J.L., Penumetcha P., Pelletier M., Sciumè G., Richard A.C., Hayes E.T., Gomez-Rodriguez J., et al. . 2014. The TNF-family cytokine TL1A promotes allergic immunopathology through group 2 innate lymphoid cells. Mucosal Immunol. 7:958–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke L.A., Groom J.R., Rankin L.C., Seillet C., Masson F., Putoczki T., and Belz G.T.. 2013. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 191:4383–4391. 10.4049/jimmunol.1301228 [DOI] [PubMed] [Google Scholar]
- Mirchandani A.S., Besnard A.G., Yip E., Scott C., Bain C.C., Cerovic V., Salmond R.J., and Liew F.Y.. 2014. Type 2 innate lymphoid cells drive CD4+ Th2 cell responses. J. Immunol. 192:2442–2448. 10.4049/jimmunol.1300974 [DOI] [PubMed] [Google Scholar]
- Mjösberg J.M., Trifari S., Crellin N.K., Peters C.P., van Drunen C.M., Piet B., Fokkens W.J., Cupedo T., and Spits H.. 2011. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat. Immunol. 12:1055–1062. 10.1038/ni.2104 [DOI] [PubMed] [Google Scholar]
- Mohapatra A., Van Dyken S.J., Schneider C., Nussbaum J.C., Liang H.E., and Locksley R.M.. 2016. Group 2 innate lymphoid cells utilize the IRF4-IL-9 module to coordinate epithelial cell maintenance of lung homeostasis. Mucosal Immunol. 9:275–286. 10.1038/mi.2015.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.B., Nussbaum J.C., Liang H.E., Van Dyken S.J., Cheng L.E., Mohapatra A., Chawla A., and Locksley R.M.. 2013. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210:535–549. 10.1084/jem.20121964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky A.B., Van Gool F., Liang H.E., Van Dyken S.J., Nussbaum J.C., Lee J., Bluestone J.A., and Locksley R.M.. 2015. Interleukin-33 and interferon-γ counter-regulate group 2 innate lymphoid cell activation during immune perturbation. Immunity. 43:161–174. 10.1016/j.immuni.2015.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaldo E., Teixeira-Alves L.G., Glatzer T., Durek P., Stervbo U., Hamann W., Babic M., Paclik D., Stölzel K., Gröne J., et al. . 2014. Human RORγt+CD34+ cells are lineage-specified progenitors of group 3 RORγt+ innate lymphoid cells. Immunity. 41:988–1000. 10.1016/j.immuni.2014.11.010 [DOI] [PubMed] [Google Scholar]
- Monticelli L.A., Sonnenberg G.F., Abt M.C., Alenghat T., Ziegler C.G., Doering T.A., Angelosanto J.M., Laidlaw B.J., Yang C.Y., Sathaliyawala T., et al. . 2011. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 12:1045–1054. 10.1038/ni.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L.A., Osborne L.C., Noti M., Tran S.V., Zaiss D.M., and Artis D.. 2015. IL-33 promotes an innate immune pathway of intestinal tissue protection dependent on amphiregulin-EGFR interactions. Proc. Natl. Acad. Sci. USA. 112:10762–10767. 10.1073/pnas.1509070112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L.A., Buck M.D., Flamar A.L., Saenz S.A., Tait Wojno E.D., Yudanin N.A., Osborne L.C., Hepworth M.R., Tran S.V., Rodewald H.R., et al. . 2016. Arginase 1 is an innate lymphoid-cell-intrinsic metabolic checkpoint controlling type 2 inflammation. Nat. Immunol. 17:656–665. 10.1038/ni.3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K., Yamada T., Tanabe M., Takeuchi T., Ikawa T., Kawamoto H., Furusawa J., Ohtani M., Fujii H., and Koyasu S.. 2010. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature. 463:540–544. 10.1038/nature08636 [DOI] [PubMed] [Google Scholar]
- Moro K., Kabata H., Tanabe M., Koga S., Takeno N., Mochizuki M., Fukunaga K., Asano K., Betsuyaku T., and Koyasu S.. 2016. Interferon and IL-27 antagonize the function of group 2 innate lymphoid cells and type 2 innate immune responses. Nat. Immunol. 17:76–86. 10.1038/ni.3309 [DOI] [PubMed] [Google Scholar]
- Mortha A., Chudnovskiy A., Hashimoto D., Bogunovic M., Spencer S.P., Belkaid Y., and Merad M.. 2014. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 343:1249288 10.1126/science.1249288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura Y., Morita H., Moro K., Nakae S., Artis D., Endo T.A., Kuroki Y., Ohara O., Koyasu S., and Kubo M.. 2014. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity. 40:758–771. 10.1016/j.immuni.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Munneke J.M., Björklund A.T., Mjösberg J.M., Garming-Legert K., Bernink J.H., Blom B., Huisman C., van Oers M.H., Spits H., Malmberg K.J., and Hazenberg M.D.. 2014. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood. 124:812–821. 10.1182/blood-2013-11-536888 [DOI] [PubMed] [Google Scholar]
- Nausch N., Appleby L.J., Sparks A.M., Midzi N., Mduluza T., and Mutapi F.. 2015. Group 2 innate lymphoid cell proportions are diminished in young helminth infected children and restored by curative anti-helminthic treatment. PLoS Negl. Trop. Dis. 9:e0003627 10.1371/journal.pntd.0003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill D.R., Wong S.H., Bellosi A., Flynn R.J., Daly M., Langford T.K., Bucks C., Kane C.M., Fallon P.G., Pannell R., et al. . 2010. Nuocytes represent a new innate effector leukocyte that mediates type-2 immunity. Nature. 464:1367–1370. 10.1038/nature08900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum J.C., Van Dyken S.J., von Moltke J., Cheng L.E., Mohapatra A., Molofsky A.B., Thornton E.E., Krummel M.F., Chawla A., Liang H.E., and Locksley R.M.. 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 502:245–248. 10.1038/nature12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oczypok E.A., Milutinovic P.S., Alcorn J.F., Khare A., Crum L.T., Manni M.L., Epperly M.W., Pawluk A.M., Ray A., and Oury T.D.. 2015. Pulmonary receptor for advanced glycation end-products promotes asthma pathogenesis through IL-33 and accumulation of group 2 innate lymphoid cells. J. Allergy Clin. Immunol. 136:747–756.e4. 10.1016/j.jaci.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard J.I., and Chawla A.. 2013. The immune system as a sensor of the metabolic state. Immunity. 38:644–654. 10.1016/j.immuni.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant C.J., Hwang Y.Y., Walker J.A., Salimi M., Wong S.H., Brewer J.M., Englezakis A., Barlow J.L., Hams E., Scanlon S.T., et al. . 2014. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4+ T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 41:283–295. 10.1016/j.immuni.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C., Uhlig H.H., and Powrie F.. 2012. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 33:289–296. 10.1016/j.it.2012.04.004 [DOI] [PubMed] [Google Scholar]
- Peebles R.S., Jr 2015. At the bedside: the emergence of group 2 innate lymphoid cells in human disease. J. Leukoc. Biol. 97:469–475. 10.1189/jlb.3BT0814-383R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.S., Han S., Xu Q., Herman M.L., Kennedy L.B., Csako G., and Bielekova B.. 2012. Inhibition of LTi cell development by CD25 blockade is associated with decreased intrathecal inflammation in multiple sclerosis. Sci. Transl. Med. 4:145ra106 10.1126/scitranslmed.3004140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettipher R., and Whittaker M.. 2012. Update on the development of antagonists of chemoattractant receptor-homologous molecule expressed on Th2 cells (CRTH2). From lead optimization to clinical proof-of-concept in asthma and allergic rhinitis. J. Med. Chem. 55:2915–2931. 10.1021/jm2013997 [DOI] [PubMed] [Google Scholar]
- Pikovskaya O., Chaix J., Rothman N.J., Collins A., Chen Y.H., Scipioni A.M., Vivier E., and Reiner S.L.. 2016. Cutting edge: Eomesodermin is sufficient to direct type 1 innate lymphocyte development into the conventional NK lineage. J. Immunol. 196:1449–1454. 10.4049/jimmunol.1502396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt J.M., Stavropoulos E., Redford P.S., Beebe A.M., Bancroft G.J., Young D.B., and O’Garra A.. 2012. Blockade of IL-10 signaling during bacillus Calmette-Guérin vaccination enhances and sustains Th1, Th17, and innate lymphoid IFN-γ and IL-17 responses and increases protection to Mycobacterium tuberculosis infection. J. Immunol. 189:4079–4087. 10.4049/jimmunol.1201061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A., and Golub R.. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12:949–958. 10.1038/ni.2105 [DOI] [PubMed] [Google Scholar]
- Price A.E., Liang H.E., Sullivan B.M., Reinhardt R.L., Eisley C.J., Erle D.J., and Locksley R.M.. 2010. Systemically dispersed innate IL-13-expressing cells in type 2 immunity. Proc. Natl. Acad. Sci. USA. 107:11489–11494. 10.1073/pnas.1003988107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak G.D., Osborne L.C., Siracusa M.C., Kim B.S., Wang K., Bayat A., Artis D., and Volk S.W.. 2016. IL-33-dependent group 2 innate lymphoid cells promote cutaneous wound healing. J. Invest. Dermatol. 136:487–496. 10.1038/JID.2015.406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A., McKenzie A.N., Carotta S., Nutt S.L., and Belz G.T.. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14:389–395. 10.1038/ni.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin L.C., Girard-Madoux M.J., Seillet C., Mielke L.A., Kerdiles Y., Fenis A., Wieduwild E., Putoczki T., Mondot S., Lantz O., et al. . 2016. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat. Immunol. 17:179–186. 10.1038/ni.3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynders A., Yessaad N., Vu Manh T.P., Dalod M., Fenis A., Aubry C., Nikitas G., Escalière B., Renauld J.C., Dussurget O., et al. . 2011. Identity, regulation and in vivo function of gut NKp46+RORγt+ and NKp46+RORγt− lymphoid cells. EMBO J. 30:2934–2947. 10.1038/emboj.2011.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roan F., Stoklasek T.A., Whalen E., Molitor J.A., Bluestone J.A., Buckner J.H., and Ziegler S.F.. 2016. CD4+ group 1 innate lymphoid cells (ILC) form a functionally distinct ILC subset that is increased in systemic sclerosis. J. Immunol. 196:2051–2062. (published erratum appears in J. Immunol. 2016. 196:3966). 10.4049/jimmunol.1501491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., and Colonna M.; Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger B., Kyle R., Yip K.H., Sumaria N., Guy T.V., Kim B.S., Mitchell A.J., Tay S.S., Jain R., Forbes-Blom E., et al. . 2013. Cutaneous immunosurveillance and regulation of inflammation by group 2 innate lymphoid cells. Nat. Immunol. 14:564–573. 10.1038/ni.2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger B., Kyle R., Tay S.S., Mitchell A.J., Bolton H.A., Guy T.V., Tan S.Y., Forbes-Blom E., Tong P.L., Köller Y., et al. . 2015. IL-2 is a critical regulator of group 2 innate lymphoid cell function during pulmonary inflammation. J. Allergy Clin. Immunol. 136:1653–1663.e7. 10.1016/j.jaci.2015.03.043 [DOI] [PubMed] [Google Scholar]
- Russi A.E., Walker-Caulfield M.E., Ebel M.E., and Brown M.A.. 2015. Cutting edge: c-Kit signaling differentially regulates type 2 innate lymphoid cell accumulation and susceptibility to central nervous system demyelination in male and female SJL mice. J. Immunol. 194:5609–5613. 10.4049/jimmunol.1500068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz S.A., Siracusa M.C., Perrigoue J.G., Spencer S.P., Urban J.F. Jr., Tocker J.E., Budelsky A.L., Kleinschek M.A., Kastelein R.A., Kambayashi T., et al. . 2010. IL25 elicits a multipotent progenitor cell population that promotes TH2 cytokine responses. Nature. 464:1362–1366. 10.1038/nature08901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz S.A., Siracusa M.C., Monticelli L.A., Ziegler C.G., Kim B.S., Brestoff J.R., Peterson L.W., Wherry E.J., Goldrath A.W., Bhandoola A., and Artis D.. 2013. IL-25 simultaneously elicits distinct populations of innate lymphoid cells and multipotent progenitor type 2 (MPPtype2) cells. J. Exp. Med. 210:1823–1837. 10.1084/jem.20122332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi M., Barlow J.L., Saunders S.P., Xue L., Gutowska-Owsiak D., Wang X., Huang L.C., Johnson D., Scanlon S.T., McKenzie A.N., et al. . 2013. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J. Exp. Med. 210:2939–2950. 10.1084/jem.20130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimi M., Xue L., Jolin H., Hardman C., Cousins D.J., McKenzie A.N., and Ogg G.S.. 2016. Group 2 innate lymphoid cells express functional NKp30 receptor inducing type 2 cytokine production. J. Immunol. 196:45–54. 10.4049/jimmunol.1501102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano T., Huang W., Hall J.A., Yang Y., Chen A., Gavzy S.J., Lee J.Y., Ziel J.W., Miraldi E.R., Domingos A.I., et al. . 2015. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector Th17 responses. Cell. 163:381–393. (published erratum appears in Cell 2016. 164:324) 10.1016/j.cell.2015.08.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanos S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., and Diefenbach A.. 2009. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22–producing NKp46+ cells. Nat. Immunol. 10:83–91. 10.1038/ni.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santori F.R., Huang P., van de Pavert S.A., Douglass E.F. Jr., Leaver D.J., Haubrich B.A., Keber R., Lorbek G., Konijn T., Rosales B.N., et al. . 2015. Identification of natural RORγ ligands that regulate the development of lymphoid cells. Cell Metab. 21:286–297. 10.1016/j.cmet.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N., Vosshenrich C.A., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. . 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29:958–970. 10.1016/j.immuni.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama N., Lesjean-Pottier S., Vieira P., Sawa S., Eberl G., Vosshenrich C.A., and Di Santo J.P.. 2010. IL-7 and IL-15 independently program the differentiation of intestinal CD3−NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207:273–280. 10.1084/jem.20092029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama N., Serafini N., Verrier T., Rekiki A., Renauld J.C., Frankel G., and Di Santo J.P.. 2014. The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity. 41:776–788. 10.1016/j.immuni.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Sawa S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., and Eberl G.. 2010. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science. 330:665–669. 10.1126/science.1194597 [DOI] [PubMed] [Google Scholar]
- Sawa S., Lochner M., Satoh-Takayama N., Dulauroy S., Bérard M., Kleinschek M., Cua D., Di Santo J.P., and Eberl G.. 2011. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat. Immunol. 12:320–326. 10.1038/ni.2002 [DOI] [PubMed] [Google Scholar]
- Scandella E., Bolinger B., Lattmann E., Miller S., Favre S., Littman D.R., Finke D., Luther S.A., Junt T., and Ludewig B.. 2008. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat. Immunol. 9:667–675. 10.1038/ni.1605 [DOI] [PubMed] [Google Scholar]
- Sciumé G., Hirahara K., Takahashi H., Laurence A., Villarino A.V., Singleton K.L., Spencer S.P., Wilhelm C., Poholek A.C., Vahedi G., et al. . 2012. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209:2331–2338. 10.1084/jem.20122097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J.A., and Grasemann H.. 2014. Arginine metabolism in asthma. Immunol. Allergy Clin. North Am. 34:767–775. 10.1016/j.iac.2014.07.007 [DOI] [PubMed] [Google Scholar]
- Seehus C.R., Aliahmad P., de la Torre B., Iliev I.D., Spurka L., Funari V.A., and Kaye J.. 2015. The development of innate lymphoid cells requires TOX-dependent generation of a common innate lymphoid cell progenitor. Nat. Immunol. 16:599–608. 10.1038/ni.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C., Rankin L.C., Groom J.R., Mielke L.A., Tellier J., Chopin M., Huntington N.D., Belz G.T., and Carotta S.. 2014. Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med. 211:1733–1740. 10.1084/jem.20140145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini N., Klein Wolterink R.G., Satoh-Takayama N., Xu W., Vosshenrich C.A., Hendriks R.W., and Di Santo J.P.. 2014. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211:199–208. 10.1084/jem.20131038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Lee J.S., Gilfillan S., Robinette M.L., Newberry R.D., Stappenbeck T.S., Mack M., Cella M., and Colonna M.. 2015. Unique and redundant functions of NKp46+ ILC3s in models of intestinal inflammation. J. Exp. Med. 212:1869–1882. 10.1084/jem.20151403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., Fouser L.A., and Artis D.. 2011a Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12:383–390. 10.1038/ni.2025 [DOI] [PubMed] [Google Scholar]
- Sonnenberg G.F., Monticelli L.A., Elloso M.M., Fouser L.A., and Artis D.. 2011b CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 34:122–134. 10.1016/j.immuni.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg G.F., Monticelli L.A., Alenghat T., Fung T.C., Hutnick N.A., Kunisawa J., Shibata N., Grunberg S., Sinha R., Zahm A.M., et al. . 2012. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 336:1321–1325. 10.1126/science.1222551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S.P., Wilhelm C., Yang Q., Hall J.A., Bouladoux N., Boyd A., Nutman T.B., Urban J.F. Jr., Wang J., Ramalingam T.R., et al. . 2014. Adaptation of innate lymphoid cells to a micronutrient deficiency promotes type 2 barrier immunity. Science. 343:432–437. 10.1126/science.1247606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N., Mebius R.E., et al. . 2013. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13:145–149. 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]
- Spooner C.J., Lesch J., Yan D., Khan A.A., Abbas A., Ramirez-Carrozzi V., Zhou M., Soriano R., Eastham-Anderson J., Diehl L., et al. . 2013. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat. Immunol. 14:1229–1236. 10.1038/ni.2743 [DOI] [PubMed] [Google Scholar]
- Tait Wojno E.D., and Artis D.. 2012. Innate lymphoid cells: balancing immunity, inflammation, and tissue repair in the intestine. Cell Host Microbe. 12:445–457. 10.1016/j.chom.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait Wojno E.D., Monticelli L.A., Tran S.V., Alenghat T., Osborne L.C., Thome J.J., Willis C., Budelsky A., Farber D.L., and Artis D.. 2015. The prostaglandin D2 receptor CRTH2 regulates accumulation of group 2 innate lymphoid cells in the inflamed lung. Mucosal Immunol. 8:1313–1323. 10.1038/mi.2015.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen M.B., Munneke J.M., Bernink J.H., Spuls P.I., Res P.C., Te Velde A., Cheuk S., Brouwer M.W., Menting S.P., Eidsmo L., et al. . 2014. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR+ ILC3 in lesional skin and blood of psoriasis patients. J. Invest. Dermatol. 134:2351–2360. 10.1038/jid.2014.146 [DOI] [PubMed] [Google Scholar]
- Tsuji M., Suzuki K., Kitamura H., Maruya M., Kinoshita K., Ivanov I.I., Itoh K., Littman D.R., and Fagarasan S.. 2008. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 29:261–271. 10.1016/j.immuni.2008.05.014 [DOI] [PubMed] [Google Scholar]
- Turner J.E., Morrison P.J., Wilhelm C., Wilson M., Ahlfors H., Renauld J.C., Panzer U., Helmby H., and Stockinger B.. 2013. IL-9–mediated survival of type 2 innate lymphoid cells promotes damage control in helminth-induced lung inflammation. J. Exp. Med. 210:2951–2965. 10.1084/jem.20130071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Pavert S.A., Ferreira M., Domingues R.G., Ribeiro H., Molenaar R., Moreira-Santos L., Almeida F.F., Ibiza S., Barbosa I., Goverse G., et al. . 2014. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 508:123–127. 10.1038/nature13158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Maele L., Carnoy C., Cayet D., Ivanov S., Porte R., Deruy E., Chabalgoity J.A., Renauld J.C., Eberl G., Benecke A.G., et al. . 2014. Activation of Type 3 innate lymphoid cells and interleukin 22 secretion in the lungs during Streptococcus pneumoniae infection. J. Infect. Dis. 210:493–503. 10.1093/infdis/jiu106 [DOI] [PubMed] [Google Scholar]
- Vonarbourg C., Mortha A., Bui V.L., Hernandez P.P., Kiss E.A., Hoyler T., Flach M., Bengsch B., Thimme R., Hölscher C., et al. . 2010. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt+ innate lymphocytes. Immunity. 33:736–751. 10.1016/j.immuni.2010.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Burg N., Chappaz S., Baerenwaldt A., Horvath E., Bose Dasgupta S., Ashok D., Pieters J., Tacchini-Cottier F., Rolink A., Acha-Orbea H., and Finke D.. 2014. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc. Natl. Acad. Sci. USA. 111:12835–12840. 10.1073/pnas.1406908111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Ji M., Liang H.E., and Locksley R.M.. 2016. Tuft-cell-derived IL-25 regulates an intestinal ILC2–epithelial response circuit. Nature. 529:221–225. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshenrich C.A., and Di Santo J.P.. 2013. Developmental programming of natural killer and innate lymphoid cells. Curr. Opin. Immunol. 25:130–138. 10.1016/j.coi.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Walford H.H., Lund S.J., Baum R.E., White A.A., Bergeron C.M., Husseman J., Bethel K.J., Scott D.R., Khorram N., Miller M., et al. . 2014. Increased ILC2s in the eosinophilic nasal polyp endotype are associated with corticosteroid responsiveness. Clin. Immunol. 155:126–135. 10.1016/j.clim.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.A., Oliphant C.J., Englezakis A., Yu Y., Clare S., Rodewald H.R., Belz G., Liu P., Fallon P.G., and McKenzie A.N.. 2015. Bcl11b is essential for group 2 innate lymphoid cell development. J. Exp. Med. 212:875–882. 10.1084/jem.20142224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers D.R., Gaspal F.M., Mackley E.C., Marriott C.L., Ross E.A., Desanti G.E., Roberts N.A., White A.J., Flores-Langarica A., McConnell F.M., et al. . 2012. Cutting edge: lymphoid tissue inducer cells maintain memory CD4 T cells within secondary lymphoid tissue. J. Immunol. 189:2094–2098. 10.4049/jimmunol.1201639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers D.R., Hepworth M.R., Wang X., Mackley E.C., Halford E.E., Dutton E.E., Marriott C.L., Brucklacher-Waldert V., Veldhoen M., Kelsen J., et al. . 2016. Transient inhibition of ROR-γt therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat. Med. 22:319–323. 10.1038/nm.4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.H., Walker J.A., Jolin H.E., Drynan L.F., Hams E., Camelo A., Barlow J.L., Neill D.R., Panova V., Koch U., et al. . 2012. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13:229–236. 10.1038/ni.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Molofsky A.B., Liang H.E., Ricardo-Gonzalez R.R., Jouihan H.A., Bando J.K., Chawla A., and Locksley R.M.. 2011. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 332:243–247. 10.1126/science.1201475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H., Keith J.W., Samilo D.W., Carter R.A., Leiner I.M., and Pamer E.G.. 2016. Innate lymphocyte/Ly6Chi monocyte crosstalk promotes Klebsiella pneumoniae clearance. Cell. 165:679–689. 10.1016/j.cell.2016.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Salimi M., Panse I., Mjösberg J.M., McKenzie A.N., Spits H., Klenerman P., and Ogg G.. 2014. Prostaglandin D2 activates group 2 innate lymphoid cells through chemoattractant receptor-homologous molecule expressed on TH2 cells. J. Allergy Clin. Immunol. 133:1184–1194.e7. 10.1016/j.jaci.2013.10.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R., Zhong C., Northrup D.L., Yu F., Bouladoux N., Spencer S., Hu G., Barron L., Sharma S., Nakayama T., et al. . 2014. The transcription factor GATA3 is critical for the development of all IL-7Rα-expressing innate lymphoid cells. Immunity. 40:378–388. 10.1016/j.immuni.2014.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Monticelli L.A., Saenz S.A., Chi A.W., Sonnenberg G.F., Tang J., De Obaldia M.E., Bailis W., Bryson J.L., Toscano K., et al. . 2013. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 38:694–704. 10.1016/j.immuni.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Li F., Harly C., Xing S., Ye L., Xia X., Wang H., Wang X., Yu S., Zhou X., et al. . 2015a TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 16:1044–1050. 10.1038/ni.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Tang T., Wei X., Yang S., and Tian Z.. 2015b Type 1 innate lymphoid cells contribute to the pathogenesis of chronic hepatitis B. Innate Immun. 21:665–673. 10.1177/1753425915586074 [DOI] [PubMed] [Google Scholar]
- Yasuda K., Muto T., Kawagoe T., Matsumoto M., Sasaki Y., Matsushita K., Taki Y., Futatsugi-Yumikura S., Tsutsui H., Ishii K.J., et al. . 2012. Contribution of IL-33-activated type II innate lymphoid cells to pulmonary eosinophilia in intestinal nematode-infected mice. Proc. Natl. Acad. Sci. USA. 109:3451–3456. 10.1073/pnas.1201042109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Pappu R., Ramirez-Carrozzi V., Ota N., Caplazi P., Zhang J., Yan D., Xu M., Lee W.P., and Grogan J.L.. 2014. TNF superfamily member TL1A elicits type 2 innate lymphoid cells at mucosal barriers. Mucosal Immunol. 7:730–740. 10.1038/mi.2013.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Wang C., Clare S., Wang J., Lee S.C., Brandt C., Burke S., Lu L., He D., Jenkins N.A., et al. . 2015. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J. Exp. Med. 212:865–874. 10.1084/jem.20142318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss M.M., Maslowski K.M., Mosconi I., Guenat N., Marsland B.J., and Harris N.L.. 2013. IL-1β suppresses innate IL-25 and IL-33 production and maintains helminth chronicity. PLoS Pathog. 9:e1003531 10.1371/journal.ppat.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaph C., Du Y., Saenz S.A., Nair M.G., Perrigoue J.G., Taylor B.C., Troy A.E., Kobuley D.E., Kastelein R.A., Cua D.J., et al. . 2008. Commensal-dependent expression of IL-25 regulates the IL-23–IL-17 axis in the intestine. J. Exp. Med. 205:2191–2198. 10.1084/jem.20080720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenewicz L.A., Yancopoulos G.D., Valenzuela D.M., Murphy A.J., Stevens S., and Flavell R.A.. 2008. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 29:947–957. 10.1016/j.immuni.2008.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., and Ouyang W.. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289. 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]
- Zhou W., Toki S., Zhang J., Goleniewksa K., Newcomb D.C., Cephus J.Y., Dulek D.E., Bloodworth M.H., Stier M.T., Polosuhkin V., et al. . 2016. Prostaglandin I2 signaling and inhibition of group 2 innate lymphoid cell responses. Am. J. Respir. Crit. Care Med. 193:31–42. 10.1164/rccm.201410-1793OC [DOI] [PMC free article] [PubMed] [Google Scholar]