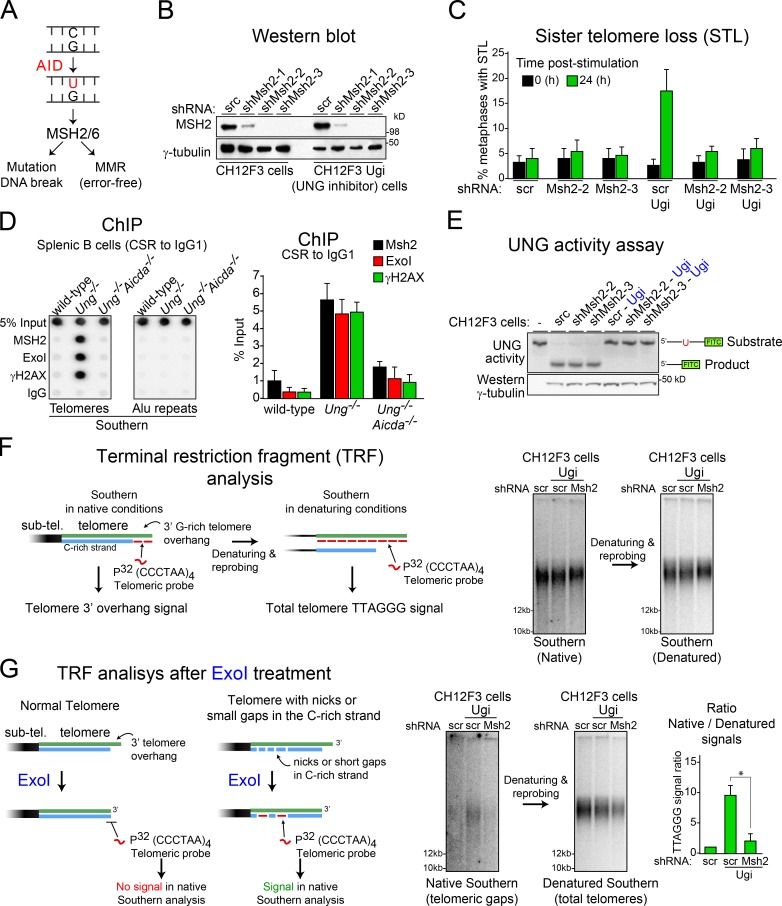
Figure 3.
Mismatch repair factors mediate AID-induced STL in Ung-deficient B cells. (A) Possible outcomes of MSH2/MSH6-initiated repair of AID-induced DNA deaminations in B cells. (B) Western blot analysis of MSH2 in CH12F3 cells expressing the indicated shRNAs. scr, scrambled. (C) Quantification of the proportion of STL per 50 metaphases in each of the different CH12F3 lines expressing or not expressing Ugi and scrambled control or two different shRNAs that deplete MSH2, before and after stimulation of CSR to IgA. Error bars represent mean + SD from at least three independent experiments. (D, left) Representative ChIP performed with the indicated antibodies in wild-type, Ung−/−, and Ung−/− Aicda−/− splenic B cells stimulated for CSR to IgG1 and analyzed by dot blotting using telomeric or Alu probes. (Right) Plot of the mean + SD dot blot signals for the telomeric probe from three independent experiments. (E) UNG activity assay using a fluorescein-labeled oligonucleotide containing a single dU, incubated with cell extracts (10 µg protein) from the indicated CH12F3 lines used in C. Substrate and product, indicated on the left, were resolved on 15% TBE-urea polyacrylamide gels. Western blot of γ-tubulin level was used as a loading control. (F) Terminal restriction fragment analysis of TTAGGG repeats in stimulated CH12F3 and CH12F3-Ugi cells expressing the indicated shRNAs via Southern blotting in native or denatured conditions. sub-tel., subtelomeric. (G, left) Diagram showing the expected outcomes after treatment of genomic DNA with exonuclease I before the TRF analyses of TTAGGG repeats. The 3′ to 5′ single-strand exonuclease activity of ExoI will remove the telomeric 3′ G-rich overhang. Therefore, the signal for single-stranded TTAGGG repeats will be lost in a TRF analysis in native conditions. However, in telomeres with short gaps or nicks in the C-rich strand, ExoI activity will expose G-rich single-stranded gaps that can be detected in a TRF analysis in native conditions. (Right) Representative Southern blots of TRF after ExoI treatment in native and denatured conditions and quantification of telomeric ssDNA/dsDNA ratio in ExoI-treated genomic DNA. Error bars represent mean + SD from at least three independent experiments. *, P < 0.003 (Student’s t test).