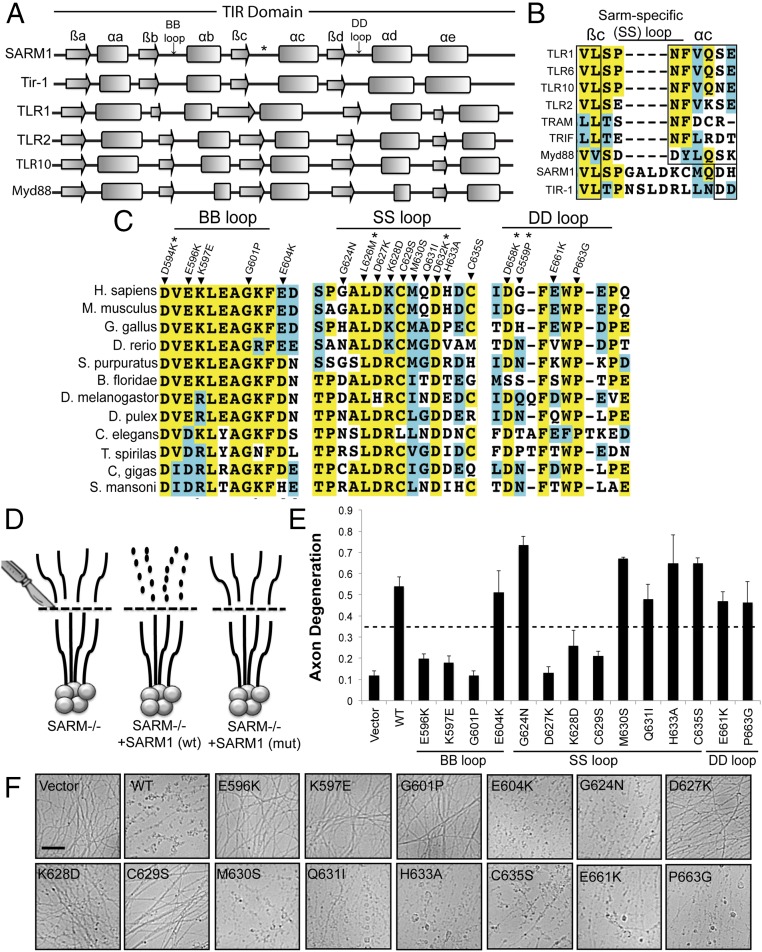
Fig. 2.
Residues within the TIR domain are required for SARM1-dependent axon degeneration. (A) Secondary structure of TIR domains from human SARM1 and C. elegans TIR-1 was predicted with three algorithms. The secondary structures of other human TIR domains are shown below. Secondary structures are derived from solved crystal structures. The β-sheets and α-helices are labeled using conventional nomenclature applied to these elements in TIR domains. (B) Extended loop region between the βc and αc elements in the TIR domain from SARM1 and TIR-1 is largely absent in other human TIR domains. We are calling this motif the SS loop. (C) Sequence alignments of TIR motifs in SARM1 orthologs. Residues highlighted in yellow are conserved. Residues highlighted in blue are amino acid changes to similar residues. Residues that not highlighted are poorly conserved. Residues chosen for mutagenesis are indicated in the alignment. *SARM1-Venus expression was very low. (D) Diagram of rescue strategy in SARM1−/− DRGs. SARM1−/− DRGs expressing full-length SARM1 restore axon degeneration, whereas SARM1 containing loss-of-function mutations (mut) does not rescue axon degeneration. (E and F) Axons from SARM1−/− DRGs expressing the indicated SARM1-Venus construct were transected with a razor blade. Axon degeneration was measured 24 h later. Degenerated axons manifest in an axon degeneration score of 0.35 or greater (indicated by dashed line). Error bars reflect ± SEM (n = 3). (Scale bar, 25 μM.)