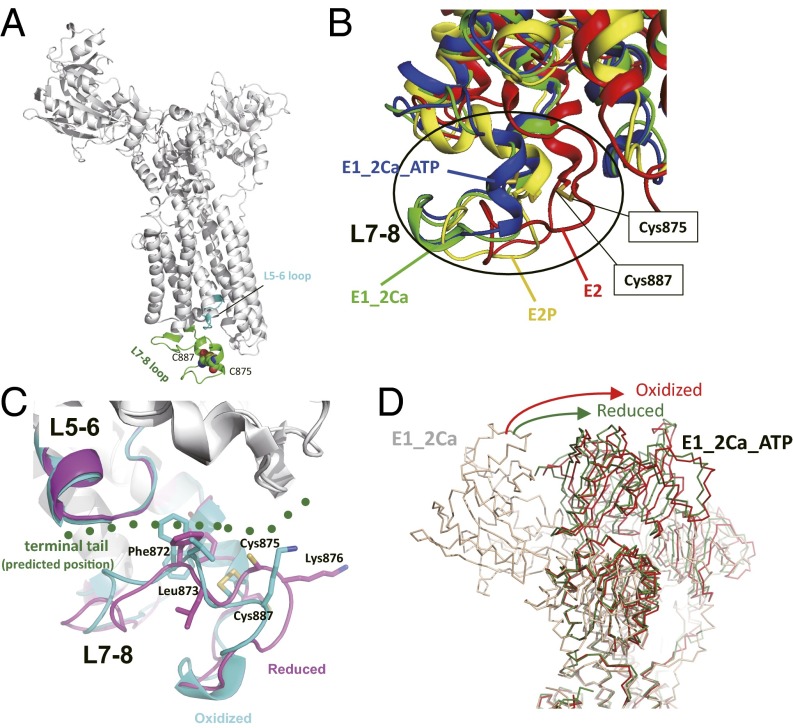
Fig. 7.
Homology models of SERCA2 provide a likely mechanism of the redox-dependent regulation of SERCA2b activity. (A) The Cys875–Cys887 pair and L5–L6 and L7–L8 loops in a SERCA2 homology model in the E1-2Ca state are highlighted. (B) Superposition of homology models of SERCA2 in the E2 (red), E1-2Ca (green), E1-2Ca-ATP (blue), and E2P (yellow) states. The homology model of SERCA2 was constructed for each state with SWISS-Model (39) using previously reported crystal structures of SERCA1a (Protein Data Bank ID: 3W5C for E2, 1SU4 for E1-2Ca, 3AR2 for E1-2Ca-ATP, and 3B9B for E2P) as templates. The transmembrane regions of these four SERCA2 models were superposed to minimize the root-mean-square deviation of their Cα atoms. Comparison of the models in different catalytic states suggests that the L7–L8 region of SERCA2 undergoes significant conformational changes during its catalytic cycle. (C) Conformational changes in the L7–L8 region upon reduction of the luminal Cys875–Cys887 disulfide bond. The homology models in the oxidized (cyan) and reduced (magenta) forms were constructed based on the crystal structures of both of the oxidized and the reduced forms of SERCA1a in the E1-2Ca-ATP state. The predicted position of the C-terminal tail of SERCA2b is shown in green dots. (D) Comparison of the relative position of the SERCA2 N domain in the E1-2Ca-ATP state. The homology models of the oxidized and reduced forms in the E1-2Ca-ATP state are shown in red and green, respectively. The model in the E1-2Ca form is shown in light brown.