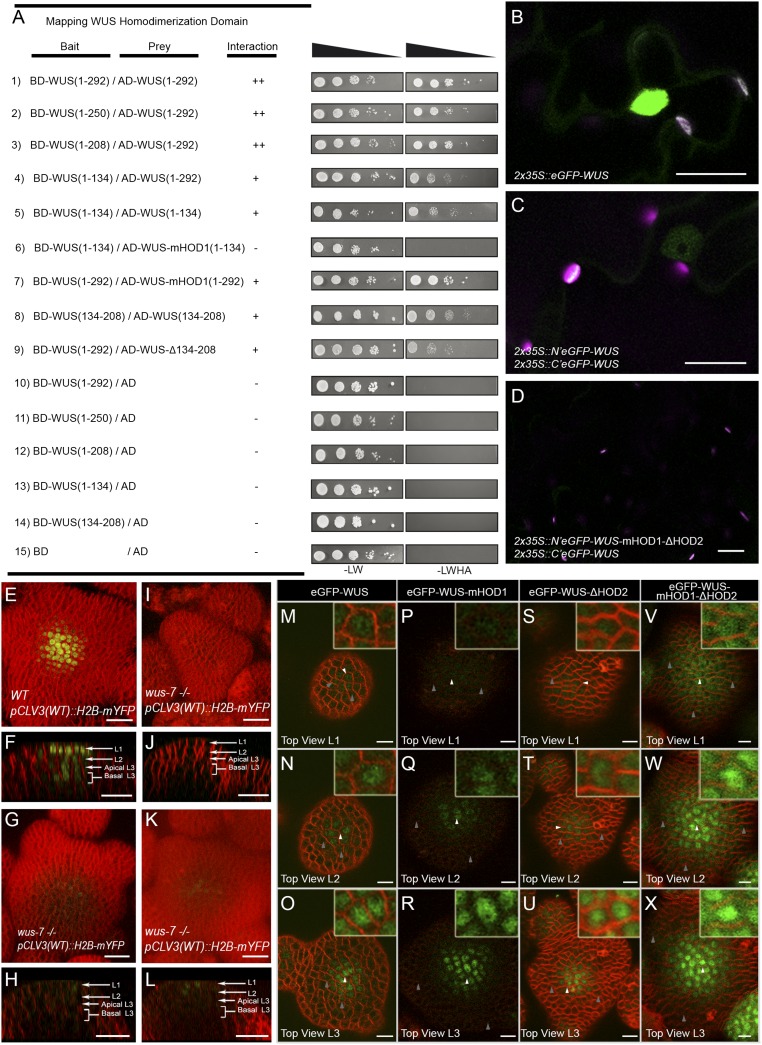
Fig. 3.
The DNA binding and dimerization of WUS restrict its spatial localization. Mapping the homodimerization domains of WUS using a yeast two-hybrid analysis (A). The break points of WUS deletions are indicated by the amino acid positions. +, stronger interaction; −, no interaction. Expression of (B) 35S::eGFP-WUS and (C) BiFC signal of WT and (D) the double homodimerization mutant in N. benthamiana leaves. Shown are 3D top views of IMs expressing WT pCLV3 (H2B-mYFP) in (E) WT and (I, G, and K) wus-7 mutant plants. A side view of E is shown in F. Side views of I, G, and K are shown in J, H, and L, respectively. H2B-mYFP images in (G, H, K, and L) were taken at three times laser intensity used in E, F, I, and J. Horizontal rows represent (M, P, S, and V) the L1 layers, (N, Q, T, and W) the L2 layers, and (O, R, U, and X) the apical L3 layers of IMs expressing (M–O) eGFP-WUS, (P–R) eGFP-WUS-mHOD1 (G77E), (S–U) eGFP-WUS-ΔHOD2 (Δ amino acid 134 to amino acid 208), and (V–X) eGFP-WUS-mHOD1(G77E)-ΔHOD2 (Δ amino acid 134 to amino acid 208) from pWUS. Insets on each panel show a higher magnification (3×) view of region identified by the white arrowheads. WUS protein detected in a broader domain is indicated by gray arrows. H2B-mYFP (yellow), eGFP (green), and FM4-64 (red). (Scale bars: B–L, 20 µm; M–X, 10 µm.)