Abstract
Bioluminescence, which living organisms such as fireflies emit light, has been studied extensively for over half a century. This intriguing reaction, having its origins in nature where glowing insects can signal things such as attraction or defense, is now widely used in biotechnology with applications of bioluminescence and chemiluminescence. Luciferase, a key enzyme in this reaction, has been well characterized; however, the enzymes involved in the biosynthetic pathway of its substrate, luciferin, remains unsolved at present. To elucidate the luciferin metabolism, we performed a de novo transcriptome analysis using larvae of the firefly species, Luciola aquatilis. Here, a comparative analysis is performed with the model coleopteran insect Tribolium casteneum to elucidate the metabolic pathways in L. aquatilis. Based on a template luciferin biosynthetic pathway, combined with a range of protein and pathway databases, and various prediction tools for functional annotation, the candidate genes, enzymes, and biochemical reactions involved in luciferin metabolism are proposed for L. aquatilis. The candidate gene expression is validated in the adult L. aquatilis using reverse transcription PCR (RT-PCR). This study provides useful information on the bio-production of luciferin in the firefly and will benefit to future applications of the valuable firefly bioluminescence system.
Keywords: Firefly bioluminescence, Functional annotation, Luciferase, RNA-seq
Introduction
The firefly is a bioluminescent beetle belonging to the Order Coleoptera, Family Lampyridae. Over 100 genera and 2,000 species of fireflies have been reported around the world both in temperate and tropical areas (McDermott, 1964; McDermott, 1966; Branham, 2010). Of these 100 genera, Photinus and Photuris from North America (Lewis & Cratsley, 2008; Faust, De Cock & Lewis, 2012; Stansbury & Moczek, 2014; Martin et al., 2015; Sander & Hall, 2015) and Pyrocoelia (Fu et al., 2006a) and Luciola (Tsutomu, Hiroki & Eiichi, 1989; Fu et al., 2006b; Oba et al., 2006; Ohtsuki et al., 2008; Oba & Kainuma, 2009; Oba et al., 2010) from Asia are the most studied, particularly their behaviors and cellular mechanisms. Bioluminescence, regarded as the most striking characteristic of fireflies, is a property generated by a chemical reaction in the photocyte cells situated in the sixth and seventh ventral segments of fireflies (Greenfield, 2001; Stanger-Hall, Lloyd & Hillis, 2007; Goh & Li, 2011). Firefly bioluminescence is catalyzed by a luciferase enzyme in the presence of O2, ATP, and Mg2+ (Deluca, 1976; Baldwin, 1996) in a two-step reaction; D-luciferin is adenylated by ATP at the luciferase active site and converted into luciferyl-adenosine monophosphate (luciferyl-AMP). This luciferyl-AMP is then oxidized and converted into excited state oxyluciferin. This excited state oxyluciferin later returns to its ground state by the emission of a visible photon, thereby generating visible light (Fraga, 2008; Naumov et al., 2009; Inouye, 2010; Pinto da Silva, Santos & Esteves da Silva, 2012).
Firefly luciferase, the key enzyme in the firefly bioluminescence reaction, is well characterized. This enzyme was first purified and crystallized in 1956 by Green and McElroy (Green & McElroy, 1956; Fraga, 2008). Later in 1985, it was cloned and expressed in Escherichia coli (De Wet et al., 1985). The structure of the luciferase from North American firefly P. pyralis was subsequently determined in 1996 (Conti, Franks & Brick, 1996). So far, firefly luciferase has been utilized in various molecular and medical studies. For instance, the firefly luciferase gene is widely used as a reporter gene in gene expression analysis (De Wet et al., 1985; Koncz et al., 1990). Firefly luciferases have been used in different applications, e.g., bioimaging (Calvo-Álvarez et al., 2015; Reimão et al., 2015), protein-protein interaction assay (Kurihara et al., 2016), immunoassay (Smirnova, Samsonova & Ugarova, 2016), and ATP quantification (Marques & Esteves da Silva, 2009). However, knowledge about the biosynthesis of the luciferase substrate, luciferin, is still lacking. To date, only the structure and chemical reactions of luciferin have been characterized (White, McCapra & Field, 1963; Fraga, 2008). The bioluminescence systems used in these applications rely solely on commercially synthesized luciferin. Many attempts to resolve the luciferin biosynthetic pathway have been performed (Okada et al., 1974; Okada, Iio & Goto, 1976; McCapra & Razavi, 1975; McCapra & Razavi, 1976; Colepicolo, Pagni & Bechara, 1988; Niwa, Nakamura & Ohmiya, 2006). Recently, Oba et al. (2013) analyzed the luciferin biosynthetic pathway by injection of isotope-labeled compounds L-cysteine, hydroquinone, and benzoquinone into an adult lantern of firefly L. lateralis. Luciferin is demonstrated to be synthesized from 1,4-hydroquinone and two endogenous L-cysteine molecules (Oba et al., 2013). The genes involved in firefly bioluminescence pathway were investigated by Viviani and colleagues in 2013. A complementary DNA (cDNA) library of Macrolampis sp2 lantern was constructed and sequenced; however, no gene product could be directly associated with luciferin biosynthesis (Viviani, Carmargo & Amaral, 2013).
For various insect species, transcriptome studies using RNA sequencing to elucidate gene networks involved in many biological pathways, e.g., olfactory mechanisms in moth (Zhang et al., 2015), visual mechanism in dragonfly (Futahashi et al., 2015), and bioluminescence mechanism in glowworm (Sharpe et al., 2015) have been done. Protein coding genes potentially involved in bioluminescent metabolism, including candidate luciferases, were identified in the New Zealand glowworm, Arachnocampa luminosa (Diptera) utilizing high-throughput sequencing technology (Sharpe et al., 2015). In Lepidoptera, the olfactory mechanisms from two pest species Helicoverpa armigera and H. assulta were studied (Zhang et al., 2015). Transcripts isolated from the antenna of the two species were sequenced using Illumina sequencing technology. They identified 133 putative chemosensory unigenes in H. armigera and 131 putative chemosensory genes in H. assulta (Zhang et al., 2015). Another example of using RNA sequencing on insect transcriptome analysis is the study of color vision opsin genes in dragonflies (Futahashi et al., 2015). This study identified 20 opsin genes in dragonflies of the Family Libellulidae (Futahashi et al., 2015). Recently, transcriptome analyses have been utilized to elucidate the opsin gene evolution in North American fireflies (Martin et al., 2015; Sander & Hall, 2015). Both RNA and genome sequencing were performed using Illumina HiSeq 2000. A total of 172 million reads were obtained from the heads of 10 firefly species. Two opsin genes were identified in their study (Sander & Hall, 2015). However, the other annotated genes derived from their transcriptome data in the study have not yet been reported.
Therefore, our study aims to reveal expressed genes in luciferin metabolism using de novo transcriptome analysis from the Illumina RNA sequencing of a Thai native firefly, L. aquatilis. Based on the transcriptome data, we used a range of protein and pathway databases combined with prediction tools to annotate the protein coding genes of L. aquatilis. Candidate genes involved in the luciferin metabolic pathway were subsequently proposed based on the studies performed by Niwa, Nakamura & Ohmiya (2006); Oba et al. (2013); Hemmati et al. (2015); Kanie et al. (2016). We validated expression of these candidate genes using reverse transcription PCR (RT-PCR) analysis in another developmental stage with the bioluminescent activity, i.e., the adult L. aquatilis. The proposed enzymes in this study provides an insight into the cryptic luciferin biosynthesis pathway in the firefly. It is worth to note that gene knockout and expression analysis are required to confirm the proposed functions of these enzymes. Prospectively, elucidation of this pathway will facilitate the development of gene reporter system, live cell imaging and other related technologies in the future.
Materials and Methods
Sample collection, RNA isolation, and RNA sequencing
L. aquatilis larvae were collected from Nakorn Ratchasrima province, Thailand. Total RNA was extracted from three specimens of bioluminescent L. aquatilis larvae. All three specimens were ground to a fine powder in liquid nitrogen with a mortar and pestle. Total RNA was extracted using TRIzol®Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The total RNA was treated with DNase I (Qiagen, Hilden, Germany) as described in the manufacturer’s protocol. Pooled RNA sample was sent for sequencing at Macrogen (South Korea) using Illumina HiSeq 2000 Sequencing System (Illumina, San Diego, CA, USA). Quality and quantity of RNA were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The mRNA was converted into a library of template molecules using the reagents provided in the Illumina®TruSeq™ RNA Sample Preparation Kit (Illumina). The poly-A containing mRNA molecules were purified using poly-T oligo-attached magnetic beads. Following purification, the mRNA is fragmented into small pieces using divalent cations under elevated temperature. The cleaved RNA fragments were converted into first strand cDNA using reverse transcriptase and random primers, followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. These cDNA fragments then went through an end repair process using an End Repair (ERP) mix. A single ‘A’ nucleotide was added to the 3′ ends of the blunt fragments to prevent them from ligating to one another during the adapter ligation reaction. Adapters were then added to the ends of the ds cDNA, preparing them for hybridization onto a flow cell. DNA Fragments were enriched by PCR and subsequently sequenced using Illumina HiSeq 2000 Sequencing System (Illumina, USA) which is able to generate paired-end read with 2 × 100 base pairs (bp) read length.
De novo transcriptome assembly
FASTQC (Version 0.11.3) was used to determine the quality of the RNA sequencing data (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were trimmed and cleaned using Trimmomatic (v. 0.32; Bolger, Lohse & Usadel, 2014). Sequences with a quality score equal to or greater than 15 and a minimum length of 36 bp were retained. After raw reads filtering, de novo assembly of transcripts data was performed using Trinity RNA-Seq assembly (release 17.07.2014; https://github.com/trinityrnaseq/trinityrnaseq/). The transcript abundance, Fragments Per Kilobase of transcript per Million mapped reads (FPKM; Trapnell et al., 2010), was calculated using Trinity based on RSEM algorithm (Li & Dewey, 2011).
Gene prediction and functional annotation of L. aquatilis
All protein-coding genes were predicted and extracted from assembled transcripts using TransDecoder (https://transdecoder.github.io/). Protein coding genes were identified via TransDecoder under the following criteria: a minimum length of 100 amino acids, a default log-likelihood score of greater than 0 and matches within the Pfam (pfam.xfam.org/) and UniProtKB/Swiss-Prot databases (web.expasy.org/docs/swiss-prot_guideline.html).
To assign protein functions, non-redundant protein (NR), and Uniprot databases were used for functional annotation. The cut-off values for each search were adjusted according to the size of the databases. An annotation with the bigger NR database was performed using a minimum amino acid sequence identity of 50% and E-value of 1E–10. Subsequently, the annotation with the smaller Uniprot database was performed using a minimum amino acid sequence under identity of 25% and E-value of 1E–05. In addition, GhostKOALA (www.kegg.jp/ghostkoala/) and BlastKOALA (www.kegg.jp/blastkoala/) tools in the metabolic pathway database, KEGG, were also used for functional annotation with a 50% identity cutoff. The annotated genes obtained from the KEGG database were categorized based on their functional roles. In addition, the protein-coding genes annotated via TransDecoder were further analyzed using KEGG taxonomic mapping.
Comparative protein sequence analysis between L. aquatilis and closely related species
The result from KEGG taxonomic mapping showed that the majority of the protein-coding genes in L. aquatilis matched to genes from various arthropod species. We further selected the most closely related species presented to assist in functional annotation. The pairwise comparison of protein sequences using BLASTP was subsequently performed between L. aquatilis and the most closely related species. The criteria for similarity searching were bidirectional best hits (BBH) with E-value of 1E–05 as a cut-off.
Identification of candidate genes, enzymes, and metabolic pathways associated with luciferin
The candidate luciferin metabolic pathway of L. aquatilis was identified based on the template luciferin pathway by Oba et al. (2013) with some modifications from different studies (Niwa, Nakamura & Ohmiya, 2006; Hemmati et al., 2015; Kanie et al., 2016). To identify all possible candidate genes and enzyme functions for assigning into each step of the biochemical reactions in the luciferin metabolic pathway of L. aquatilis, protein sequences of L. aquatilis obtained from this study were blasted against three different protein and pathway databases, NR, KEGG, and Uniprot using BLASTP. The protein sequences that showed the best hits under the highest identity to one of the NR, KEGG, or Uniprot databases were retained. The candidate genes that have functions associated with luciferin were then selected. Moreover, literature search and manual curation were also performed to identify all possible putative enzymes involved in each step of the luciferin metabolic pathway.
Validation of candidate gene expression using RT-PCR
To validate the expression of these candidate genes in the L. aquatilis, a different developmental stage, i.e., adult stage, which is also bioluminescent was selected for RT-PCR analysis. Total RNA was extracted from an adult L. aquatilis using the same method as previously described. The first strand DNA was amplified using RevertAid first strand cDNA (Thermo Scientific, USA). Reverse transcription reaction was performed using 2 μg of total RNA and oligo-dT primer as described in the user manual. PCR was then performed using Phire Hot Start II DNA Polymerase (Thermo Scientific, USA). The PCR reaction included 2 μL of the reverse transcriptase reaction mix, 1X Phire Reaction Buffer, 200 μM of each dNTPs, 0.5 μM of each primer (File S1), and Phire Hot Start II DNA Polymerase 1 Unit. The PCR reaction was performed under the following conditions: initial denaturation for 5 min at 98°C, 35 amplification cycles of denaturation for 40 s at 98°C, annealing for 30 s at 50°C, extension for 1 min at 72°C, and final extension for 5 min at 72°C. The amplified products were visualized by 1% agarose gel electrophoresis.
Results and Discussion
Transcriptome assembly of L. aquatilis
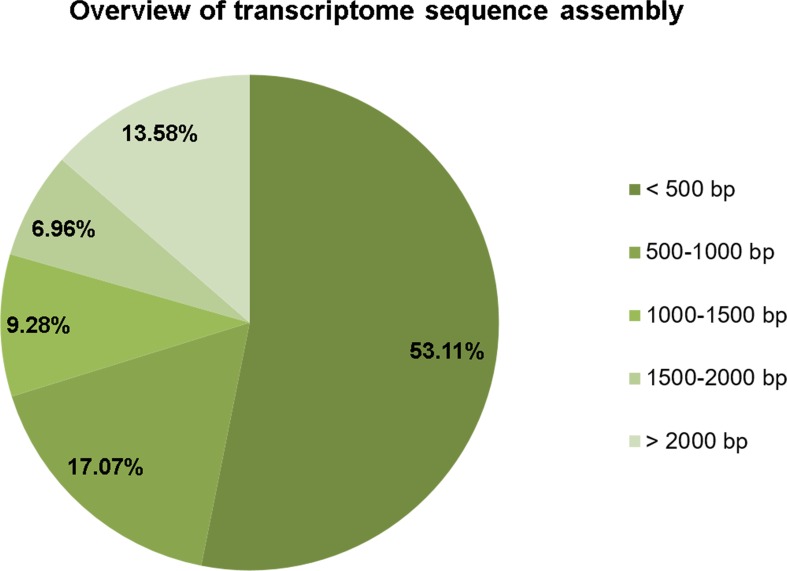
The RNA extracted from the bioluminescent L. aquatilis larvae was sequenced using the Illumina HiSeq 2000 platform. A total of 63,533,268 raw reads with an average read length of 101 bp were obtained. After adapter trimming and removing of contaminating sequences and low-quality sequences, a total of 62,481,222 trimmed reads were retained. These trimmed reads were assembled into high-quality contigs with a total length of 38,873,002 bp and a total number of 39,730 contigs. The number of contigs without isoforms was 33,070. The maximum length of a contig was 26,786 bp, and the minimum length of a contig was 201 bp with an average length of 978.43 bp; an N50 of 1,889 bp were obtained (Table 1). The size distribution of contigs (Fig. 1) demonstrated that 21,102 contigs were <500 bp (53.11%), 6,780 contigs were 500–1,000 bp (17.07%), 3,686 contigs were 1,000–1,500 bp (9.28%), 2,767 contigs were 1,500–2,000 bp (6.96%), and 5,395 contigs were >2,000 bp (13.58%). The clean reads of L. aqualtilis larvae in this study were deposited in the NCBI SRA database under the accession number SRX1605859.
Table 1. Overview of the transcriptome.
The transcriptome data of L. aquatilis larvae was obtained from Illumina HiSeq 2000 platform.
| Info | All transcript contigs (only longest isoform per ‘GENE’) |
|---|---|
| Total raw reads | 63,533,268 |
| Total raw nucleotide | 6,416,860,068 |
| Total clean reads | 62,481,222 |
| Total clean nucleotides | 6,228,699,940 |
| Q20 percentage | 99.20% |
| GC percentage | 42.03% |
| Total trinity ‘genes’ | 39,730 (33,070) |
| Total trinity transcripts | 39,730 (33,070) |
| Maximum length of contigs | 26,786 (26,786) |
| Minimum length of contigs | 201 (201) |
| Median length of contigs | 451 (384) |
| Mean length of contigs | 978.43 (833.32) |
| N50 of unigenes | 1,889 (1,630) |
| Total assembled bases | 38,873,002 (27,557,931) |
Figure 1. Size distribution of contigs.
The size distribution of contigs demonstrated that the majority of the sequences was <500 bps.
Functional annotation of L. aquatilis
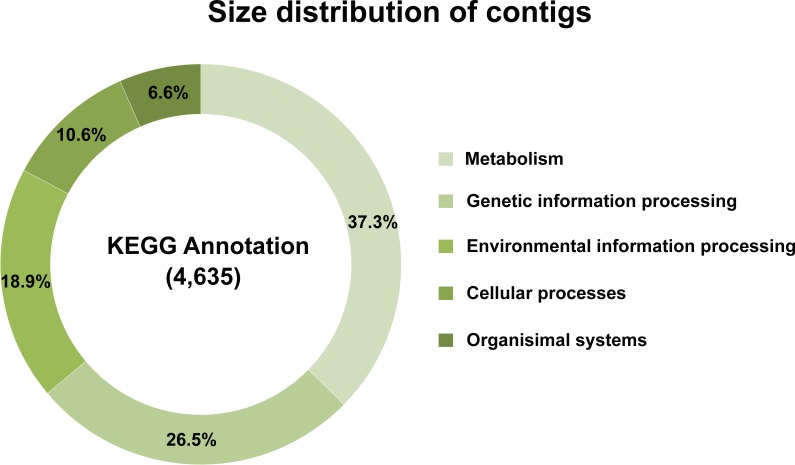
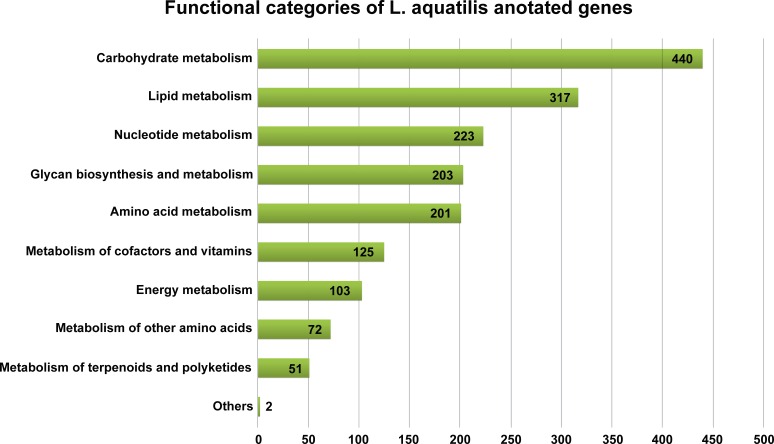
Of 39,730 assembled contigs, 19,761 protein-coding genes were identified using TransDecoder. These genes were subsequently annotated using three databases, KEGG, NR, and Uniprot, and the best match sequences from each of the databases were retained. A total of 14,025 (70.97%), 14,855 (75.17%), and 4,976 (25.18%) annotated protein-coding genes were obtained from NR, Uniprot, and KEGG, respectively (File S2). To further elucidate the functional and pathway association, the KEGG database was used (Kanehisa et al., 2011). Of the 4,976 KEGG annotated genes, a total of 4,653 genes were grouped into five functional categories, i.e., metabolism (1,737 genes, 37.3%), genetic information processing (1,233 genes, 26.5%), environmental information processing (881 genes, 18.9%), cellular processes (494 genes, 10.6%), and organismal systems (308 genes, 6.6%) (Fig. 2). From these results, the majority of predicted protein-coding genes from L. aquatilis were involved in metabolic functions. The genes in this functional category were further distributed into ten subcategories (Fig. 3). The genes associated in carbohydrate metabolism, lipid metabolism, and amino acid metabolism showed the highest numbers. This can be explained by the fact that genes in these three categories are involved in basic processes in living cells and are highly conserved and well-characterized pathways across the animal kingdom (Peregrín-Alvarez, Sanford & Parkinson, 2009). These results are consistent with transcriptome studies in other coleopteran insects, e.g., Colaphellus bowringi (Tan et al., 2015) and Tomicus yunnanensis (Zhu, Zhao & Yang, 2012). In contrast, the metabolism of the terpenoids and polyketides categories contained the least gene member, which is expected, as these secondary metabolites could be specific to each species. These compounds play several important roles in ecological interactions and also evolutionary aspects (Pankewitz & Hilker, 2008). Different groups of beetles have been reported to produce different defensive compounds. In the family Coccinellidae (ladybugs), many compounds, e.g., coccinelline from Coccinella septempunctata (Tursch et al., 1971), hippodamine from Hippodamia convergens (Tursch et al., 1972), and propyleine from Propylaea quatuordecimpunctata (Tursch, Daloze & Hootele, 1972) were identified. Both larva and adult leaf beetles (family Chrysomelidae) are also known to produce defensive compounds such as isoaxazolinone glucoside 5 and its 3-nitropropionate esters 6 to prevent them from natural enemies (Pauls et al., 2016). In the larvae of carabid beetle, Chlaenius cordicollis, various defensive compounds, e.g., methylhydroquinone, toluquinone 2,3-dimethylquinone were detected (Holliday, Mattingly & Holliday, 2015). Due to these diverse yet species-specific metabolites found among groups of insect, when the database searches were performed, only a small number of genes were annotated as being involved in the metabolism of terpenoids and polyketides in L. aquatilis.
Figure 2. Overview of KEGG annotation.
A total of 4,653 genes were grouped into five functional categories with the majority in the metabolism category.
Figure 3. Metabolism functional categories of L. aquatilis annotated genes.
The genes categorized in the metabolism category were further categorized into ten subcategories with the majority in the carbohydrate metabolism.
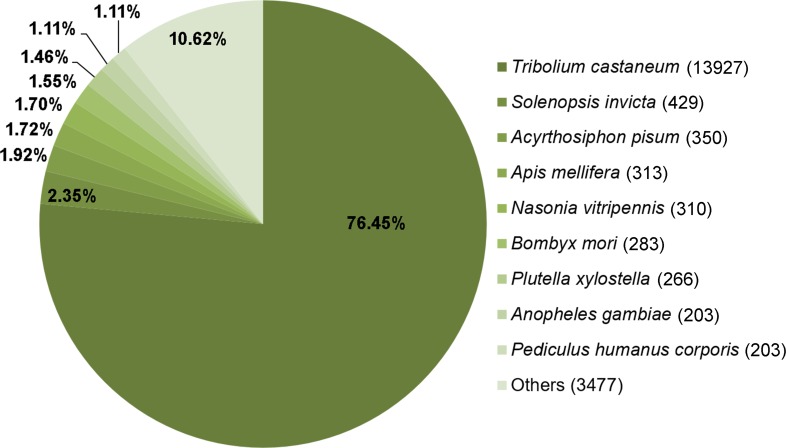
To identify the orthologues of these annotated protein-coding genes in other taxa, KEGG taxonomic mapping was performed using the 19,761 genes obtained from TransDecoder. The majority of these sequences (>80%) matched the genes found in other arthropods. Of 13,927 genes, 70.48% observably matched the genes found in Tribolium castaneum, followed by Solenopsis invicta (429 sequences; 2.17%), Acyrthosiphon pisum (350 sequences; 1.77%), Apis mellifera (313 sequences; 1.58%), Bombyx mori (283 sequences; 1.43%), and the others (4,149 sequences; 21.01%) (Fig. 4). This result verifies the data obtained in this study as T. casteneum is the closest group of insect with whole genome data available for comparison. Therefore, it is not surprising that L. aquatilis would have the most orthologues with T. castaneum. Based on this, we selected to use the T. castaneum genome as the reference for further pathway mapping analysis.
Figure 4. Comparative analysis of L. aquatilis KEGG-annotated genes with other arthropods.
KEGG taxonomic mapping was performed using the 19,761 genes obtained from TransDecoder. The majority of these sequences matched the genes found in Tribolium castaneum.
Comparative protein sequence analysis between L. aquatilis and T. castaneum
Based on KEGG taxonomic mapping, T. castaneum showed the highest homologues with L. aquatilis (76.45%). Although this T. castaneum is not bioluminescence, it is a model organism of Coleopteran insect, providing the most extensive list of proteins with which to compare; therefore, we selected this species for the comparative analysis. A comparative protein sequence analysis between these two species was then performed to identify conserved genes and their functions. The bidirectional best hits (BBH) analysis was performed between L. aquatilis (19,761 genes) and T. castaneum (18,076 genes). As a result, a total of 8,020 conserved genes were identified (File S3). These conserved genes are mostly involved in common biological pathways found in most insects, such as growth and development, lipid metabolism, and energy metabolism, e.g., juvenile hormone epoxide hydrolase, fatty acid hydroxylase, and V-type proton ATPase, respectively (File S3).
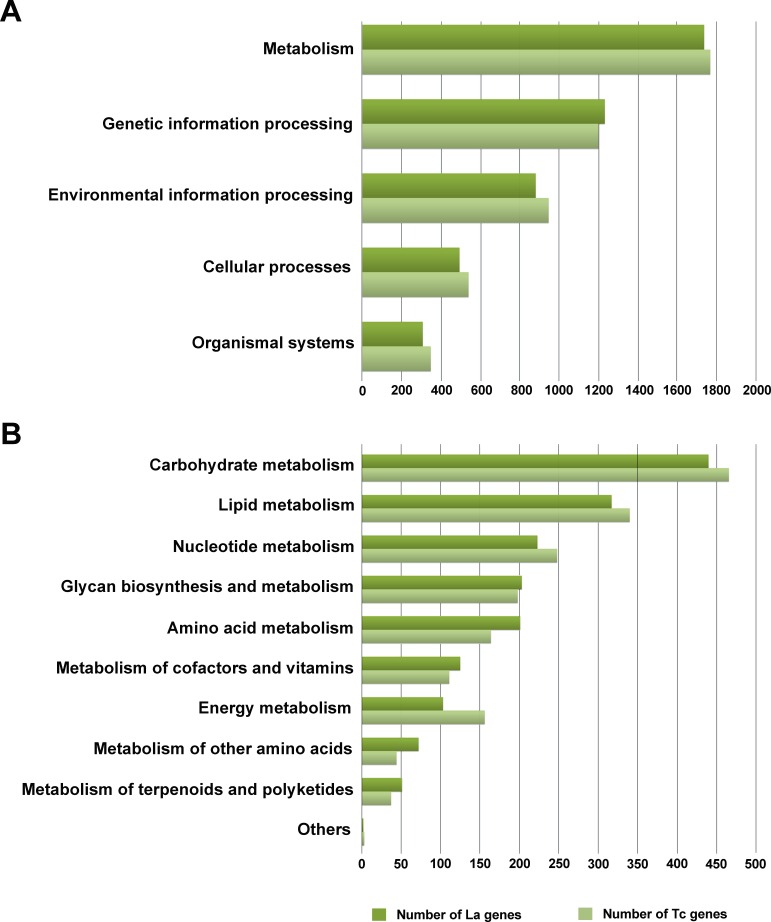
Assessing gene functional distribution in KEGG between L. aquatilis (4,653 genes) and T. castaneum (4,803 genes), similar trends were found as illustrated in Fig. 5, the highest number of genes was found in the metabolism category. Interestingly, the numbers of genes in energy metabolism, nucleotide metabolism, lipid metabolism, and carbohydrate metabolism were higher in T. castaneum than in L. aquatilis. In contrast, the numbers of genes in the metabolism of cofactors and vitamins, amino acid metabolism, metabolism of terpenoids and polyketides, metabolism of other amino acids, and glycan biosynthesis and metabolism were higher in L. aquatilis than in T. castaneum. It is worth noting that our transcriptome data was obtained only from the larvae of the firefly, but the data set from T. castaneum included annotated protein data from the genome that represents the proteins in both adult and larval stages (ftp://ftp.ncbi.nih.gov/genomes/Tribolium_castaneum/protein/). Not surprising, the genes involved in energy metabolism are likely to be higher in number in T. castaneum. In many insects, the processes during larval stages are mainly involved in food and energy storage for intense growth, while reproduction tends to be the main focus during the adult stage (Arnold, Cassey & White, 2016). These results coincided with feeding behaviors of both species. T. castaneum has an extremely carbohydrate-rich diet. It is considered a pest of storage grains and cereal products (Perkin, Elpidina & Oppert, 2016). In contrast, the larvae of L. aquatilis have a protein-rich diet as they consume only aquatic snails during this larval stage (Thancharoen, Ballantyne & Branham, 2007). On the contrary, genes in the metabolism of terpenoids and polyketides were higher in the L. aquatilis than in the T. castaneum, suggesting the complexity of the terpenoids and polyketides pathways in the firefly. Apart from the juvenile hormones that are present in both T. castaneum and L. aquatilis, not many other terpenoids and polyketides were reported in the T. castaneum eventhough it is a model beetle, and the genes have been well characterized. Several quinone derivatives were reported to be defensive compounds in the T. castaneum (Unruh, Xu & Kramer, 1998; Villaverde, Juárez & Mijailovsky, 2007). Another compound of terpenoids and polyketides found in T. castaneum was 4,8-Dimethyldecanal (4,8-DMD), an aggregation pheromone produced by male T. castaneum (Kim, Matsuyama & Suzuki, 2005). In fireflies, defensive steroidal pyrones (lucibufagins) released via a defensive behavior called reflexed-bleeding were detected in many genera including Photinus, Ellychnia, and Lampyris (González, Hare & Eisner, 1999; Tyler et al., 2008; Mosey et al., 2015). Another compound, a defensive betaine, N-methylquinolinium 2-carboxylate, was found in the adults and female larvae of Photuris (González et al., 1999; Trice, Tyler & Day, 2004). Furthermore, over 10 novel steroids from Lucidota atra were detected using capillary NMR spectroscopy (Gronquist et al., 2005). Although a direct relationship between these defensive compounds and bioluminescence reaction has never been reported, bioluminescence in the firefly is often used for defensive signaling to warn predators of its non-palatability and toxicity (Trice, Tyler & Day, 2004; Fu et al., 2007).
Figure 5. Comparative protein sequence analysis between L. aquatilis and T. casteneum.
A pairwise comparison of putative gene sequences between L. aquatilis and T. castaneum demonstrated a similar functional distribution trend, with the highest number of genes in the metabolism category (A). The genes in this metabolism category were further compared in the sub-category level (B).
Identification of candidate genes involved in luciferin metabolic pathway
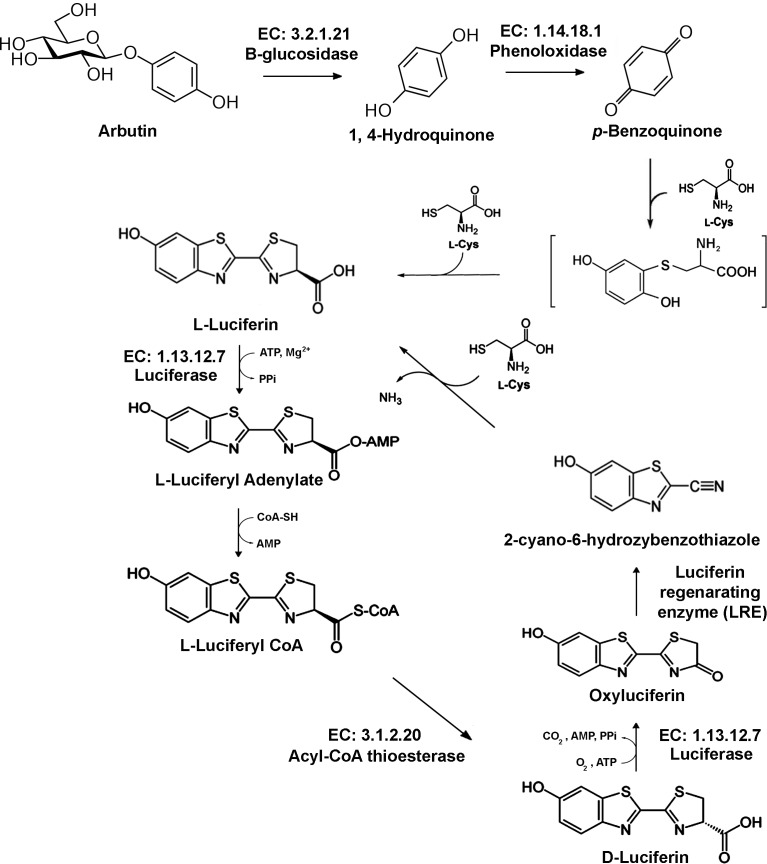
A total of 11 candidate genes involved in the luciferin metabolic pathway were identified in the L. aquatilis transcriptome (Table 2 and Fig. 6). These genes were selected from protein sequences that matched one of the NR, KEGG, or Uniprot databases. These 11 genes, were annotated as putative β-glucosidase enzymes (EC: 3.2.1.21), phenoloxidase (EC: 1.14.18.1), luciferase (EC: 1.13.12.7), thioesterase (EC: 3.1.2.20), and luciferin regenerating enzyme (LRE). Most of the candidates obtained for this study used integration of NR and Uniprot, whereas none of the candidates were identified using KEGG. This result indicated that the KEGG database contains limited information about protein functions associated with bioluminescence, whether for fireflies or any other bioluminescent insects (Kanehisa et al., 2016), in comparison with the other two databases.
Table 2. List of candidate enzymes involved in the luciferin metabolic pathway.
A total of 11 candidate genes involved in the luciferin metabolic pathway were identified in the L. aquatilis transcriptome data. An elongation factor 1-alpha was also identified to be used as a control in the RT-PCR experiment.
| EC number | Protein name | Transcript ID | FPKM | Protein ID | Functional description | Accession | Database | Amino acid sequence identity (%) | E-value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| NR | KEGG | Uniprot | |||||||||
| EC: 3.2.1.21 | B-glucosidases | c14185_g1_i1 | 10.85 | c14185_g1_i1| m.15351 | Aryl-phospho-beta-D-glucosidase BglA [Bacillus subtilis (strain 168)] | Q17PP1 | ✓a | ✓ | 44.95 | 2E–58 | |
| c11559_g1_i1 | 1.3 | c11559_g1_i1| m.9194 | Cytosolic beta-glucosidase [Cavia porcellus] | P97265 | ✓a | ✓ | 41.99 | 9E–99 | |||
| EC:1.14.18.1 | Phenol oxidases | c14353_g1_i1 | 230.43 | c14353_g1_i1| m.15909 | Phenoloxidase 2 [Drosophila melanogaster] | Q9V521 | ✓a | ✓ | 57.37 | 0 | |
| c14376_g1_i1 | 320.14 | c14376_g1_i1| m.15966 | Phenoloxidase 2 [Drosophila melanogaster] | Q9V521 | ✓ | ✓ | 62.26 | 0 | |||
| EC: 1.13.12.7 | Luciferases | c10041_g1_i1 | 112.72 | c10041_g1_i1| m.6619 | Luciferase [Luciola lateralis] | Q01158.1 | ✓ | ✓ | 83.67 | 0 | |
| c13054_g1_i1 | 65.83 | c13054_g1_i1| m.12283 | Firefly luciferase [Luciola cruciata] | BAJ41368.1 | ✓ | ✓ | 79.04 | 0 | |||
| EC: 3.1.2.20 | Thioesterases | c9513_g1_i1 | 9.32 | c9513_g1_i1| m.5804 | Acyl-coenzyme A thioesterase 13 [Saimiri boliviensis boliviensis] | XP_003927347.1 | ✓ | ✓a | 87.06 | 0 | |
| c13177_g1_i1 | 9.7 | c13177_g1_i1| m.12638 | Acyl-coenzyme A thioesterase 10, mitochondrial [Tribolium castaneum] | XP_973760.2 | ✓ | ✓a | 60.86 | 7E–179 | |||
| N/A | Luciferin regenerating enzyme (LRE) | c10156_g1_i1 | 55.04 | c10156_g1_i1| m.6798 | Luciferin regenerating enzyme [Lampyris turkestanicus] | ADK55065.1 | ✓ | ✓a | 78.18 | 0 | |
| c12106_g1_i1 | 24.61 | c12106_g1_i1| m.10324 | Luciferin-regenerating enzyme [Luciola cruciata] | BAB85479.1 | ✓ | ✓a | 74.43 | 2E–170 | |||
| c8279_g1_i1 | 0.68 | c8279_g1_i1| m.4268 | Luciferin-regenerating enzyme [Luciola cruciata] | BAB85479.1 | ✓ | ✓a | 56.11 | 4E–43 | |||
| N/A | Elongation factor 1-alpha | c11516_g1_i1 | 4.3 | c11516_g1_i1| m.9114 | Elongation factor 1-alpha [Tribolium castaneum] | XP_966355.1 | ✓ | ✓ | ✓ | 94.24 | 0 |
Notes.
Predicted gene name in the database differs from what is presented in the subject column (File S2).
Figure 6. Proposed luciferin metabolic pathway.
A total of 11 candidate genes involved in the luciferin metabolic pathway were identified in the L. aquatilis transcriptome data (adapted from Niwa, Nakamura & Ohmiya, 2006; Oba et al., 2013; Hemmati et al., 2015; Kanie et al., 2016).
Proposed luciferin metabolism of L. aquatilis
The luciferin biosynthesis is thought to be generated from 1,4-hydroquinone (Oba et al., 2013; Fig. 6). This 1,4-hydroquinone is proposed to be stored as arbutin in the firefly lantern. Only arbutin, but not 1,4-hydroquinone, was detected in the adult firefly lantern by HPLC, suggesting that 1,4-hydroquinone was immediately oxidized to 1,4-benzoquinone to produce luciferin (Oba et al., 2013). This 1,4-hydroquinone is hydrolyzed from arbutin by arbutin hydrolysis enzymes, i.e., glucosidases (Reinhard et al., 2002; Oba et al., 2013). In this study, we identified two putative β-glucosidase enzymes, c14185 (FPKM: 10.85) and c11559 (FPKM: 1.3) from the transcriptome data. However, hydroquinone has other functions in the developmental pathway of arthropods as it can also be used to crosslink proteins in the cuticle (Shear, 2015). This suggests the role of hydroquinone in L. aquatilis larvae could be as a precursor of both the luciferin pathway and cuticle production.
The next step in luciferin metabolism is an oxidation of 1,4-hydroquinone into 1,4-benzoquinone (Fig. 6). The incorporation efficiency of the [D4]-benzoquinone into firefly luciferin was higher than that of [D6]-hydroquinone, indicating that 1,4-hydroquinone may convert to 1,4-benzoquinone in the biosynthesis of luciferin in the firefly lantern (Oba et al., 2013). The transcriptome analysis of the T. castaneum odoriferous defensive stink gland revealed candidate enzymes, i.e., glucosidases, phenol oxidases, and peroxidases, that are involved in the production of quinones (Li et al., 2013). The substrate (1, 4-hydroquinone) and the product (p-benzoquinone) were searched in the T. castaneum KEGG pathway. The pathway that was identified to convert 1,4-hydroquinone into p-benzoquinone was tca00740 (riboflavin metabolism). In fireflies, p-benzoquinone is a precursor for luciferin biosynthesis (Oba et al., 2013) and it could possibly be synthesized using phenol oxidases. In this study, two candidate genes encoding for phenol oxidases, c14353 (FPKM: 230.43) and c14376 (FPKM: 320.14), were identified. Benzoquinones are toxic compounds reported to be produced in many arthropods including beetles as a defensive mechanism (Ibarra & Blair, 2013; Rocha et al., 2013). In cockchafer, benzoquinone is used to attract mates and protect the larvae from pathogenic bacteria and fungi (Ruther et al., 2001; Ruther, Podsiadlowski & Hilker, 2001). However, the defensive compounds in fireflies were reported to be steroid pyrones called lucibufagins (Eisner et al., 1978; Meinwald, Wiemer & Eisner, 1979; Eisner et al., 1997) and betaine N-methylquinolinium 2-carboxylate (González et al., 1999). The lucibufagins are produced by the adult of the North American firefly genus Photinus (Eisner et al., 1978; Eisner et al., 1997). Interestingly, the adult fireflies of the genus Photuris acquire these compounds from Photinus fireflies through consumption (Eisner et al., 1997; Faust, De Cock & Lewis, 2012). These defensive compounds are released when the fireflies are disturbed through the chemical defensive behavior called “reflexed-bleeding” (Blum & Sannasi, 1974; Eisner et al., 1997). This behavior is also observed in other firefly genera, e.g., Pyrocoelia (Wang et al., 2007) and Asymmetricata (A Sriboonlert, pers. obs., 2013). In the larvae of many genera including Lampyris, Luciola, and Nyctophila, pleural defensive organs have been identified which secrete a repellent substance used as a defensive mechanism (Trice, Tyler & Day, 2004). In aquatic firefly larvae, Luciola leii, a closely related species of L. aquatilis it is reported to produce two volatile terpenoids: terpinolene and γ-terpinene from thoracic and abdominal glands as repellent compounds (Fu et al., 2006b; Fu et al., 2007). From these findings, benzoquinone is unlikely to be used directly in fireflies as a defensive compound, and instead is likely solely used for the production of luciferin. Nonetheless, it is possible that these quinone substances could also have indirect benefits as defensive compounds against pathogenic bacteria and predators.
After p-benzoquinone is obtained, it is converted into L-luciferin in the presence of L- cysteine (Fig. 6). This reaction has been proven to occur nonenzymatically (Kanie et al., 2016). L-luciferin is demonstrated to be generated from p-benzoquinone and cysteine in various neutral buffers without any enzymes (Kanie et al., 2016). Nonetheless, L-luciferin was demonstrated to act as a D-luciferin antagonist in bioluminescence (Lembert, 1996; Niwa, Nakamura & Ohmiya, 2006; Nakamura et al., 2005). The chirality of luciferin is also important in the bioluminescence reaction (Niwa, Nakamura & Ohmiya, 2006). In the firefly lantern, L-luciferin was produced from L-cysteine. However, the substrate for luciferase in firefly bioluminescence was reported to be D-luciferin, whereas L-luciferin acts as an inhibitor of the bioluminescence reaction (Seliger et al., 1961; Lembert, 1996; Niwa, Nakamura & Ohmiya, 2006; Inouye, 2010). Endogenous luciferin from the adult fireflies was detected in both D- and L-forms, with more of the D-form than the L-form (Niwa, Nakamura & Ohmiya, 2006; Oba et al., 2013). With evidence from the incorporation study and the measurement of D- and L-luciferin levels in different developmental stages of firefly, this suggests that that the L-luciferin is a biosynthetic intermediate of D-luciferin (Niwa, Nakamura & Ohmiya, 2006; Oba et al., 2013). The conversion of L-luciferin to D-luciferin is demonstrated to be an enzymatic reaction (Niwa, Nakamura & Ohmiya, 2006) with ATP, Mg2+, and CoA suggesting the function of CoA-thioesterase hydrolysis of D-luciferyl CoA to yield D-luciferin (Niwa, Nakamura & Ohmiya, 2006; Hunt et al., 2006; Niwa, Nakamura & Ohmiya, 2006; Niwa, Nakajima & Ohmiya, 2010). However, Inouye (2010) suggest the racemization between D-LH2-AMP and L-LH2 AMP might not occur in the luciferase molecule, but in the solution non-enzymatically after releasing luciferyl, forming adenylate from the luciferase molecule. In this study, two acyl-CoA thioesterases, c9513 (FPKM: 9.32) and c13177 (FPKM: 9.7), corresponding to EC: 3.1.2.20 were identified. Relatively low expression of this enzyme could be due to the low accumulation of D-luciferin at the larval stage. In L. cruciata, D-luciferin was detected at the highest concentration in the adult stage (Niwa, Nakamura & Ohmiya, 2006).
The next step of the luciferin metabolic pathway is where the actual bioluminescence occurs (Fig. 6). Luciferase is crucial for the bioluminescence reaction. Firefly luciferase (EC. 1.13.12.7) oxidizes the luciferin substrate with the presence of cofactors Mg2+, O2, and ATP to produce oxyluciferin and emit yellow-green light (Inouye, 2010). In this study, two candidate luciferases, c10041 (FPKM: 112.72) and c13054 (FPKM: 65.83), were identified. One of these luciferases, c10041, demonstrated the highest FPKM of 112.72 and has the highest homology to L. lateralis luciferase, with 83.67% identity. Another luciferase enzyme, c13054, with FPKM of 65.83, also has high homology to the L. cruciata luciferase (79.04%). In addition, five putative luciferases were identified in this study, i.e., c12163 (FPKM: 17), c14996 (FPKM: 5.31), c18617 (FPKM: 1.09), c13390 (FPKM: 3.73), and c14833 (FPKM: 5.58) (File S2), with much lower identity scores (<45%) and FPKM. These luciferases may have other functions not involving bioluminescence. Previous studies report only one luciferase enzyme in each species that is responsible for the bioluminescence reaction (Viviani et al., 2004). However, a recent study demonstrates a luciferase isotype LcLuc2 in Luciola cruciata (Oba et al., 2010). Both LcLuc1 and LcLuc2 show luminescence activity and fatty acyl-CoA synthetic activity (Tsutomu, Hiroki & Eiichi, 1989; Oba et al., 2010). In addition, luciferase paralogs (LcLL1 and LcLL2) are also identified in the same species (Oba et al., 2006). However, neither the LcLL1 nor LcLL2 show enzymatic activity (Oba et al., 2006). Apart from the bioluminescence reaction, luciferase is found to catalyze fatty acyl-CoA synthesis from fatty acids in the presence of ATP, Mg2+, and CoA. The luciferase enzyme is hypothesized to have evolved from fatty acyl-CoA synthetase (Inouye, 2010). Moreover, the fatty acyl-CoA synthetase enzyme from non-luminous insects can be converted into luciferase by site-directed mutagenesis (Inouye, 2010), suggesting the other luciferase candidates identified in this study may actually be fatty acyl-CoA synthetase enzyme.
The last step in the luciferin metabolic pathway is the recycling of oxyluciferin. Oxyluciferin is reported to be recycled into 2-cyano-6-hydrobenzothiazole (CHBT) by luciferin regenerating enzyme (LRE; Fig. 6) (Gomi & Kajiyama, 2001; Gomi, Hirokawa & Kajiyama, 2002; Day, Tisi & Bailey, 2004; Niwa, Nakamura & Ohmiya, 2006; Marques & da Silva, 2009; Emamzadeh et al., 2010; Inouye, 2010; Hemmati et al., 2015). The CHBT then combines with L-cysteine and is converted into L-luciferin (Fig. 6). This step is reported to occur without enzymes (Okada et al., 1974; Gomi & Kajiyama, 2001; Day, Tisi & Bailey, 2004). Incorporation experiments demonstrate incorporations of 2-cyano-6-hydrobenzothiazole (CHBT), and D- and L- cysteine into D-luciferin in fireflies (McCapra & Razavi, 1976; Okada, Iio & Goto, 1976; Day, Tisi & Bailey, 2004; Niwa, Nakamura & Ohmiya, 2006). Both D- and L- forms of luciferin are found in the firefly at a different ratio at different stages of development (Lembert, 1996; Niwa, Nakamura & Ohmiya, 2006). In the presence of LRE and D-cysteine, an increase of bioluminescence is observed, but in the absence of LRE, bioluminescence only appears for nine minutes (Gomi & Kajiyama, 2001; Gomi, Hirokawa & Kajiyama, 2002; Hemmati et al., 2015). In this study, we identified three putative LREs, c10156 (FPKM: 55.04), c12106 (FPKM: 24.61), and c8279 (FPKM: 0.68). The c10156 DNA sequence was very similar to the T-LRE from Lampyris turkestanicus (Alipour et al., 2004), with 78.18% amino acid sequence identity. The other two LREs, c12106 and c8279, show highest similarity to the LRE from Luciola cruciata (Gomi, Hirokawa & Kajiyama, 2002) with 74.43% and 56.11% amino acid sequence identity, respectively. This is the first report of multiple transcripts of LRE identified in one species of firefly. However, the role of these three LREs in L. aquatilis remains to be confirmed.
Validation of candidate gene expression using RT-PCR
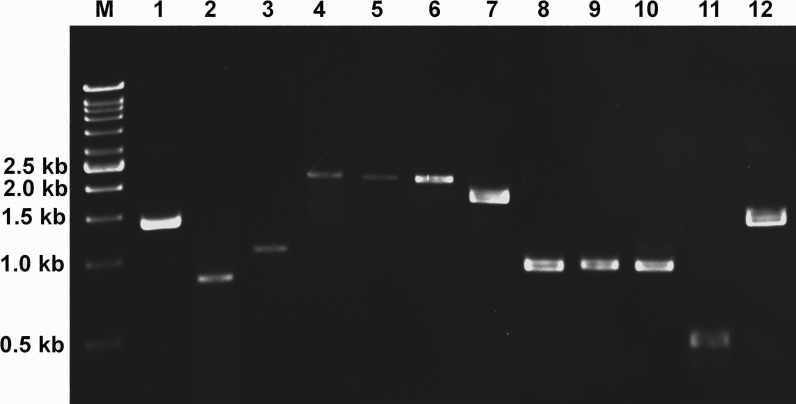
The expression of 11 candidate genes identified in this study was verified using RT-PCR (Fig. 7). This validation was performed using a different developmental stage from the RNA-seq experiment to confirm the expression of these candidate genes in L. aquatilis. All candidate genes involved in the luciferin metabolic pathway are expected to be expressed in both larval and adult stages as fireflies at these two developmental stages are bioluminescence. Of seven candidate luciferase genes, only the two with the highest identity with known firefly luciferases were selected for RT-PCR analysis. An elongation factor 1-alpha (EF1α) gene (c11516; FPKM: 4.3) was selected as an internal control because it was presented in all three databases. It is not a common reference gene, but it was used because it is one of the most stable reference genes in another coleopteran, C. bowringi (Tan et al., 2015). On the other hand, more common reference genes, e.g., tubulin and GAPDH, were shown to be less stable (Tan et al., 2015). The results show that the 11 candidate genes are expressed, verifying the transcriptome data analyzed in this study. Nonetheless, further experiments e.g., gene knockout and gene expression analysis are required to prove the function of these candidate genes in each step of luciferin biosynthesis.
Figure 7. Gene expression analysis of candidate genes using reverse transcription PCR (RT-PCR).
Expression of candidate genes in luciferin metabolic pathway was analyzed by RT-PCR. Elongation factor 1-alpha (lane 1) was used as a control. M: DNA ladder (1 Kb), 1: c11516_g1_i1 (Elongation factor 1-alpha, 1,383 bp), 2: c14185_g1_i1 (B-glucosidase, 819 bp), 3: c11559_g1_i1 (B-glucosidase, 1,049 bp), 4: c14353_g1_i1 (Phenoloxidase, 2,058 bp), 5: c14376_g1_i1 (Phenoloxidase, 2,046 bp), 6: c13054_g1_i1 (Luciferase, 2,022 bp), 7: c10041_g1_i1 (Luciferase, 1,635 bp), 8: c10156_g1_i1 (Luciferin regenerating enzyme, 921 bp), 9: c12106_g1_i1 (Luciferin regenerating enzyme, 930 bp), 10: c8279_g1_i1 (Luciferin regenerating enzyme, 927 bp), 11: c9513_g1_i1 (Thioesterase, 450 bp), 12: c13177_g1_i1 (Thioesterase, 1,374 bp).
Conclusions
In this study, we identify candidate genes involved in the firefly bioluminescence reaction from transcriptome data of bioluminescent L. aquatilis larvae. Here, we proposed a list of enzymes involved in the firefly luciferin metabolic pathway. The expression of these enzyme-encoding genes is demonstrated in the adult stage of the firefly to confirm our transcriptome results. Although candidate enzymes of the luciferin biosynthetic pathway have been identified in this study, the actual function of these enzymes still need to be verified. Gene knockout will be performed in the fireflies to confirm the functions of these candidate genes by using the recent genome editing technology. Moreover, gene expression analyses will also be performed to confirm the involvement of these enzymes in the luciferin metabolic pathway. By elucidating the luciferin biosynthetic pathway, the development of firefly bioluminescence applications will be extended. Currently, applications using the firefly bioluminescence reporter system primarily rely on commercially synthesized luciferin to generate luminescence. Many applications can benefit from the elucidation of luciferin biosynthesis. Autoluminescence of modified organisms is ideal for this reporter system. Other bioluminescence systems, e.g., bacterial lux that can be seen without any light sources, have also been used in similar applications. Recently, a group of researchers generated an autoluminescent plant from a bacterial lux system (Krichevsky et al., 2010). However, one of the main problems of the bacterial lux system is the bioluminescence intensity especially in the in vivo bioluminescence assay (Mudge, Lewis-Henderson & Birch, 1996). In contrast, firefly LUC system was proven to be more efficient detectable by luminometer assays in the intact tissues of plants (Mudge, Lewis-Henderson & Birch, 1996). Prior attempts to create firefly bioluminescent plants were first examined in 1986 (Ow et al., 1986), but as the luciferin biosynthesis process was still mysterious, the plants could only glow in contact with applied luciferin substrate. By understanding firefly luciferin metabolic pathways, we can develop a self-sustainable system without having to constantly apply the luciferin substrate. This also has potential to be used in live cell imaging and related technologies in the future.
Supplemental Information
Acknowledgments
We would like to thank Mr. Preecha Patumcharoenpol and Miss Sarintip Nguantad for assisting in the gene prediction process and functional annotation, respectively.
Funding Statement
This work was funded by Kasetsart University Research and Development Intitute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Wanwipa Vongsangnak and Pramote Chumnanpuen analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Ajaraporn Sriboonlert conceived and designed the experiments, performed the experiments, contributed reagents/materials/analysis tools, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
GenBank (SRX1605859).
Data Availability
The following information was supplied regarding data availability:
The raw data has been supplied as Supplementary File.
References
- Alipour et al. (2004).Alipour BS, Hosseinkhani S, Nikkhah M, Naderi-Manesh H, Chaichi MJ, Osaloo SK. Molecular cloning, sequence analysis, and expression of a cDNA encoding the luciferase from the glow-worm, Lampyris turkestanicus. Biochemical and Biophysical Research Communications. 2004;325:215–222. doi: 10.1016/j.bbrc.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Arnold, Cassey & White (2016).Arnold PA, Cassey P, White CR. Maturity matters for movement and metabolic rate: trait dynamics across the early adult life of red flour beetles. Animal Behaviour. 2016;111:181–188. doi: 10.1016/j.anbehav.2015.10.023. [DOI] [Google Scholar]
- Baldwin (1996).Baldwin TO. Firefly luciferase: the structure is known, but the mystery remains. Structure. 1996;4:223–228. doi: 10.1016/S0969-2126(96)00026-3. [DOI] [PubMed] [Google Scholar]
- Blum & Sannasi (1974).Blum MS, Sannasi A. Reflex bleeding in the lampyrid Photinus pyralis: defensive function. Journal of Insect Physiology. 1974;20:451–460. doi: 10.1016/0022-1910(74)90153-X. [DOI] [Google Scholar]
- Bolger, Lohse & Usadel (2014).Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branham (2010).Branham M. Handbook of zoology. Vol. 4. Walter de Gruyter GmbH & Co.; Berlin: 2010. Lampyridae Latreille, 1817; pp. 141–145. [Google Scholar]
- Calvo-Álvarez et al. (2015).Calvo-Álvarez E, Álvarez-Velilla R, Fernández-Prada C, Balaña-Fouce R, Reguera RM. Trypanosomatids see the light: recent advances in bioimaging research. Drug Discovery Today. 2015;20:114–121. doi: 10.1016/j.drudis.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Colepicolo, Pagni & Bechara (1988).Colepicolo P, Pagni D, Bechara EJH. Luciferin biosynthesis in larval Pyrearinus termitilluminans (Coleoptera: Elateridae) Comparative Biochemistry and Physiology Part B. 1988;91:143–147. doi: 10.1016/0305-0491(88)90126-5. [DOI] [Google Scholar]
- Conti, Franks & Brick (1996).Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/S0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- Day, Tisi & Bailey (2004).Day JC, Tisi LC, Bailey MJ. Evolution of beetle bioluminescence: the origin of beetle luciferin. Luminescence. 2004;19:8–20. doi: 10.1002/bio.749. [DOI] [PubMed] [Google Scholar]
- De Wet et al. (1985).De Wet JR, Wood KV, Helinski DR, Deluca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca (1976).Deluca M. Firefly luciferase. In: Meister A, editor. Advances in enzymology and related areas of molecular biology. Vol. 44. John Wiley & Sons; New York: 1976. pp. 37–68. [DOI] [PubMed] [Google Scholar]
- Eisner et al. (1997).Eisner T, Goetz MA, Hill DE, Smedley SR, Meinwald J. Firefly “femmes fatales” acquire defensive steroids (lucibufagins) from their firefly prey. Proceedings of the National Academy of Sciences of the United States of America of the United States of America. 1997;94:9723–9728. doi: 10.1073/pnas.94.18.9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisner et al. (1978).Eisner T, Wiemer DF, Haynes LW, Meinwald J. Lucibufagins: defensive steroids from the fireflies Photinus ignitus and P. marginellus (Coleoptera: Lampyridae) Proceedings of the National Academy of Sciences of the United States of America. 1978;75:905–908. doi: 10.1073/pnas.75.2.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamzadeh et al. (2010).Emamzadeh R, Hosseinkhani S, Hemati R, Sadeghizadeh M. RACE-based amplification of cDNA and expression of a luciferin-regenerating enzyme (LRE): an attempt towards persistent bioluminescent signal. Enzyme and Microbial Technology. 2010;47:159–165. doi: 10.1016/j.enzmictec.2010.05.008. [DOI] [Google Scholar]
- Faust, Cock & Lewis (2012).Faust L, De Cock R, Lewis S. Thieves in the night: kleptoparasitism by fireflies in the genus Photuris Dejean (Coleoptera: Lampyridae) The Coleopterists Bulletin. 2012;66:1–6. doi: 10.2307/41413688. [DOI] [Google Scholar]
- Fraga (2008).Fraga H. Firefly luminescence: a historical perspective and recent developments. Photochemical & Photobiological Sciences. 2008;7:146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- Fu et al. (2006a).Fu X, Nobuyoshi O, Meyer-Rochow V, Wang Y, Lei C. Reflex-bleeding in the firefly Pyrocoelia pectoralis (Coleoptera: Lampyridae): morphological basis and possible function. The Coleopterists Bulletin. 2006a;60:207–215. doi: 10.1649/892.1. [DOI] [Google Scholar]
- Fu et al. (2006b).Fu X, Nobuyoshi O, Vencl FV, Lei C. Life cycle and behaviour of the aquatic firefly Luciola leii (Coleoptera: Lampyridae) from Mainland China. The Canadian Entomologist. 2006b;138:860–870. doi: 10.4039/n05-093. [DOI] [Google Scholar]
- Fu et al. (2007).Fu X, Vencl FV, Nobuyoshi O, Meyer-Rochow VB, Lei C, Zhang Z. Structure and function of the eversible glands of the aquatic firefly Luciola leii (Coleoptera: Lampyridae) Chemoecology. 2007;17:117–124. doi: 10.1007/s00049-007-0370-3. [DOI] [Google Scholar]
- Futahashi et al. (2015).Futahashi R, Kawahara-Miki R, Kinoshita M, Yoshitake K, Yajima S, Arikawa K, Fukatsu T. Extraordinary diversity of visual opsin genes in dragonflies. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1247–E1256. doi: 10.1073/pnas.1424670112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh & Li (2011).Goh K-S, Li C-W. A photocytes-associated fatty acid-binding protein from the light organ of adult Taiwanese firefly, Luciola cerata. PLoS ONE. 2011;6:e29576. doi: 10.1371/journal.pone.0029576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi, Hirokawa & Kajiyama (2002).Gomi K, Hirokawa K, Kajiyama N. Molecular cloning and expression of the cDNAs encoding luciferin-regenerating enzyme from Luciola cruciata and Luciola lateralis. Gene. 2002;294:157–166. doi: 10.1016/S0378-1119(02)00764-3. [DOI] [PubMed] [Google Scholar]
- Gomi & Kajiyama (2001).Gomi K, Kajiyama N. Oxyluciferin, a luminescence product of firefly luciferase, is enzymatically regenerated into luciferin. Journal of Biological Chemistry. 2001;276:36508–36513. doi: 10.1074/jbc.M105528200. [DOI] [PubMed] [Google Scholar]
- González, Hare & Eisner (1999).González A, Hare JF, Eisner T. Chemical egg defense in Photuris firefly “femmes fatales”. Chemoecology. 1999;9:177–185. doi: 10.1007/s000490050051. [DOI] [Google Scholar]
- González et al. (1999).González A, Schroeder F, Meinwald J, Eisner T. N-Methylquinolinium 2-carboxylate, a defensive betaine from Photuris versicolor fireflies 1. Journal of Natural Products. 1999;62:378–380. doi: 10.1021/np980400o. [DOI] [PubMed] [Google Scholar]
- Green & Mcelroy (1956).Green AA, McElroy W. Crystalline firefly luciferase. Biochimica et Biophysica ACTA. 1956;20:170–176. doi: 10.1016/0006-3002(56)90275-X. [DOI] [PubMed] [Google Scholar]
- Greenfield (2001).Greenfield MD. Missing link in firefly bioluminescence revealed: NO regulation of photocyte respiration. BioEssays. 2001;23:992–995. doi: 10.1002/bies.1144. [DOI] [PubMed] [Google Scholar]
- Gronquist et al. (2005).Gronquist M, Meinwald J, Eisner T, Schroeder FC. Exploring uncharted terrain in nature’s structure space using capillary NMR spectroscopy: 13 steroids from 50 fireflies. Journal of the American Chemical Society. 2005;127:10810–10811. doi: 10.1021/ja053617v. [DOI] [PubMed] [Google Scholar]
- Hemmati et al. (2015).Hemmati R, Hosseinkhani S, Sajedi RH, Azad T, Tashakor A, Bakhtiari N, Ataei F. Luciferin-regenerating enzyme mediates firefly luciferase activation through direct effects of D-cysteine on luciferase structure and activity. Photochemistry and Photobiology. 2015;91:828–836. doi: 10.1111/php.12430. [DOI] [PubMed] [Google Scholar]
- Holliday, Mattingly & Holliday (2015).Holliday AE, Mattingly TM, Holliday NJ. Defensive secretions of larvae of a carabid beetle. Physiological Entomology. 2015;40:131–137. doi: 10.1111/phen.12096. [DOI] [Google Scholar]
- Hunt et al. (2006).Hunt MC, Rautanen A, Westin MA, Svensson LT, Alexson SE. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters shows that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. The FASEB Journal. 2006;20:1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- Ibarra & Blair (2013).Ibarra Y, Blair NT. Benzoquinone reveals a cysteine-dependent desensitization mechanism of TRPA1. Molecular Pharmacology. 2013;83:1120–1132. doi: 10.1124/mol.112.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye (2010).Inouye S. Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cellular and Molecular Life Sciences. 2010;67:387–404. doi: 10.1007/s00018-009-0170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa et al. (2011).Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2011;40(Database issue):D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa et al. (2016).Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanie et al. (2016).Kanie S, Nishikawa T, Ojika M, Oba Y. One-pot non-enzymatic formation of firefly luciferin in a neutral buffer from p-benzoquinone and cysteine. Scientific Reports. 2016;6 doi: 10.1038/srep24794. Article 24794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Matsuyama & Suzuki (2005).Kim J, Matsuyama S, Suzuki T. 4, 8-Dimethyldecanal, the aggregation pheromone of Tribolium castaneum, is biosynthesized through the fatty acid pathway. Journal of Chemical Ecology. 2005;31:1381–1400. doi: 10.1007/s10886-005-5292-3. [DOI] [PubMed] [Google Scholar]
- Koncz et al. (1990).Koncz C, Langridge WH, Olsson O, Schell J, Szalay AA. Bacterial and firefly luciferase genes in transgenic plants: advantages and disadvantages of a reporter gene. Developmental Genetics. 1990;11:224–232. doi: 10.1002/dvg.1020110308. [DOI] [Google Scholar]
- Krichevsky et al. (2010).Krichevsky A, Meyers B, Vainstein A, Maliga P, Citovsky V. Autoluminescent plants. PLoS ONE. 2010;5:e15461. doi: 10.1371/journal.pone.0015461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara et al. (2016).Kurihara M, Ohmuro-Matsuyama Y, Ayabe K, Yamashita T, Yamaji H, Ueda H. Ultra sensitive firefly luciferase-based protein-protein interaction assay (FlimPIA) attained by hinge region engineering and optimized reaction conditions. Biotechnology Journal. 2016;11:91–99. doi: 10.1002/biot.201500189. [DOI] [PubMed] [Google Scholar]
- Lembert (1996).Lembert N. Firefly luciferase can use L-luciferin to produce light. Biochemical Journal. 1996;317:273–277. doi: 10.1042/bj3170273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis & Cratsley (2008).Lewis SM, Cratsley CK. Flash signal evolution, mate choice, and predation in fireflies. Annual Review of Entomology. 2008;53:293–321. doi: 10.1146/annurev.ento.53.103106.093346. [DOI] [PubMed] [Google Scholar]
- Li & Dewey (2011).Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2013).Li J, Lehmann S, Weißbecker B, Naharros IO, Schütz S, Joop G, Wimmer EA. Odoriferous defensive stink gland transcriptome to identify novel genes necessary for quinone synthesis in the red flour beetle, Tribolium castaneum. PLoS Genetics. 2013;9:e1003596. doi: 10.1371/journal.pgen.1003596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques & da Silva (2009).Marques SM, Esteves da Silva JC. Firefly bioluminescence: a mechanistic approach of luciferase catalyzed reactions. IUBMB Life. 2009;61:6–17. doi: 10.1002/iub.134. [DOI] [PubMed] [Google Scholar]
- Martin et al. (2015).Martin GJ, Lord NP, Branham MA, Bybee SM. Review of the firefly visual system (Coleoptera: Lampyridae) and evolution of the opsin genes underlying color vision. Organisms Diversity & Evolution. 2015;15:513–526. doi: 10.1007/s13127-015-0212-z. [DOI] [Google Scholar]
- McCapra & Razavi (1975).McCapra F, Razavi Z. A model for firefly luciferin biosynthesis. Journal of the Chemical Society, Chemical Communications. 1975;2:42–43. doi: 10.1039/C3975000042B. [DOI] [Google Scholar]
- McCapra & Razavi (1976).McCapra F, Razavi Z. Biosynthesis of luciferin in Pyrophorus pellucens. Journal of the Chemical Society, Chemical Communications. 1976;5:153–154. doi: 10.1039/C39760000153. [DOI] [Google Scholar]
- McDermott (1964).McDermott FA. The taxonomy of the Lampyridae (Coleoptera) Transactions of the American Entomological Society. 1964;90:1–72. [Google Scholar]
- McDermott (1966).McDermott FA. Lampyridae. Coleopterorum Catalogus. 1966;Supplement, Part 9:1–149. [Google Scholar]
- Meinwald, Wiemer & Eisner (1979).Meinwald J, Wiemer DF, Eisner T. Lucibufagins. 2. Esters of 12-oxo-2. beta., 5. beta., 11. alpha.-trihydroxybufalin, the major defensive steroids of the firefly Photinus pyralis (Coleoptera: Lampyridae) Journal of the American Chemical Society. 1979;101:3055–3060. doi: 10.1021/ja00505a037. [DOI] [Google Scholar]
- Mosey et al. (2015).Mosey C, Gaber M, Ahmed Z, Risteen R, Smedley S, Deyrup ST. Chemical investigation of winter fireflies (Ellychnia corrusca) reveals lucibufagins. Planta Medica. 2015;81:PI1. doi: 10.1055/s-0035-1556262. [DOI] [Google Scholar]
- Mudge, Lewis-Henderson & Birch (1996).Mudge SR, Lewis-Henderson WR, Birch RG. Comparison of vibrio and firefly luciferases as reporter gene systems for use in bacteria and plants. Australian Journal of Plant Physiology. 1996;23:75–83. doi: 10.1071/PP9960075. [DOI] [Google Scholar]
- Nakamura et al. (2005).Nakamura M, Maki S, Amano Y, Ohkita Y, Niwa K, Hirano T, Ohmiya Y, Niwa H. Firefly luciferase exhibits bimodal action depending on the luciferin chirality. Biochemical and Biophysical Research Communications. 2005;331:471–475. doi: 10.1016/j.bbrc.2005.03.202. [DOI] [PubMed] [Google Scholar]
- Naumov et al. (2009).Naumov P, Ozawa Y, Ohkubo K, Fukuzumi S. Structure and spectroscopy of oxyluciferin, the light emitter of the firefly bioluminescence. Journal of the American Chemical Society. 2009;131:11590–11605. doi: 10.1021/ja904309q. [DOI] [PubMed] [Google Scholar]
- Niwa, Nakajima & Ohmiya (2010).Niwa K, Nakajima Y, Ohmiya Y. Applications of luciferin biosynthesis: bioluminescence assays for l-cysteine and luciferase. Analytical Biochemistry. 2010;396:316–318. doi: 10.1016/j.ab.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Niwa, Nakamura & Ohmiya (2006).Niwa K, Nakamura M, Ohmiya Y. Stereoisomeric bio-inversion key to biosynthesis of firefly D-luciferin. FEBS Letters. 2006;580:5283–5287. doi: 10.1016/j.febslet.2006.08.073. [DOI] [PubMed] [Google Scholar]
- Oba & Kainuma (2009).Oba Y, Kainuma T. Diel changes in the expression of long wavelength-sensitive and ultraviolet-sensitive opsin genes in the Japanese firefly, Luciola cruciata. Gene. 2009;436:66–70. doi: 10.1016/j.gene.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Oba et al. (2010).Oba Y, Mori N, Yoshida M, Inouye S. Identification and characterization of a luciferase isotype in the Japanese firefly, Luciola cruciata, involving in the dim glow of firefly eggs. Biochemistry. 2010;49:10788–10795. doi: 10.1021/bi1016342. [DOI] [PubMed] [Google Scholar]
- Oba et al. (2006).Oba Y, Sato M, Ohta Y, Inouye S. Identification of paralogous genes of firefly luciferase in the Japanese firefly, Luciola cruciata. Gene. 2006;368:53–60. doi: 10.1016/j.gene.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Oba et al. (2013).Oba Y, Yoshida N, Kanie S, Ojika M, Inouye S. Biosynthesis of firefly luciferin in adult lantern: decarboxylation of L-cysteine is a key step for benzothiazole ring formation in firefly luciferin synthesis. PLoS ONE. 2013;8:e84023. doi: 10.1371/journal.pone.0084023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuki et al. (2008).Ohtsuki H, Yokoyama J, Ohba N, Ohmiya Y, Kawata M. Nitric oxide synthase (NOS) in the Japanese fireflies Luciola lateralis and Luciola cruciata. Archives of Insect Biochemistry and Physiology. 2008;69:176–188. doi: 10.1002/arch.20275. [DOI] [PubMed] [Google Scholar]
- Okada, Iio & Goto (1976).Okada K, Iio H, Goto T. Biosynthesis of firefly luciferin. Probable formation of benzothiazole from p-benzoquinone and cysteine. Journal of the Chemical Society, Chemical Communications. 1976;1:32–32. doi: 10.1039/C39760000032. [DOI] [Google Scholar]
- Okada et al. (1974).Okada K, Iio H, Kubota I, Goto T. Firefly bioluminescence III. Conversion of Oxyluciferin to luciferin in firefly. Tetrahedron Letters. 1974;15:2771–2774. doi: 10.1016/S0040-4039(01)91738-1. [DOI] [Google Scholar]
- Ow et al. (1986).Ow DW, De Wet JR, Helinski DR, Howell SH, Wood KV, Deluca M. Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science. 1986;234:856–859. doi: 10.1126/science.234.4778.856. [DOI] [PubMed] [Google Scholar]
- Pankewitz & Hilker (2008).Pankewitz F, Hilker M. Polyketides in insects: ecological role of these widespread chemicals and evolutionary aspects of their biogenesis. Biological Reviews of the Cambridge Philosophical Society. 2008;83:209–226. doi: 10.1111/j.1469-185X.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- Pauls et al. (2016).Pauls G, Becker T, Rahfeld P, Gretscher RR, Paetz C, Pasteel J, Von Reuss SH, Burse A, Boland W. Two defensive lines in juvenile leaf beetles; esters of 3-nitropropionic acid in the hemolymph and aposematic warning. Journal of Chemical Ecology. 2016;42:240–248. doi: 10.1007/s10886-016-0684-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrín-Alvarez, Sanford & Parkinson (2009).Peregrín-Alvarez JM, Sanford C, Parkinson J. The conservation and evolutionary modularity of metabolism. Genome Biology. 2009;10 doi: 10.1186/gb-2009-10-6-r63. Article R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkin, Elpidina & Oppert (2016).Perkin L, Elpidina EN, Oppert B. Expression patterns of cysteine peptidase genes across the Tribolium castaneum life cycle provide clues to biological function. PeerJ. 2016;4:e1581. doi: 10.7717/peerj.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto da silva, Santos & Esteves da Silva (2012).Pinto da Silva LS, Santos AJM, Esteves da Silva JC. Efficient firefly chemi/bioluminescence: evidence for chemiexcitation resulting from the decomposition of a neutral firefly dioxetanone molecule. The Journal of Physical Chemistry A. 2012;117:94–100. doi: 10.1021/jp311711p. [DOI] [PubMed] [Google Scholar]
- Reimão et al. (2015).Reimão JQ, Oliveira JC, Trinconi CT, Cotrim PC, Coelho AC, Uliana SRB. Generation of luciferase-expressing Leishmania infantum chagasi and assessment of miltefosine efficacy in infected hamsters through bioimaging. PLoS Neglected Tropical Diseases. 2015;9:e0003556. doi: 10.1371/journal.pntd.0003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard et al. (2002).Reinhard J, Lacey MJ, Ibarra F, Schroeder FC, Kaib M, Lenz M. Hydroquinone: a general phagostimulating pheromone in termites. Journal of Chemical Ecology. 2002;28:1–14. doi: 10.1023/A:1013554100310. [DOI] [PubMed] [Google Scholar]
- Rocha et al. (2013).Rocha DF, Wouters FC, Zampieri DS, Brocksom TJ, Machado G, Marsaioli AJ. Harvestman phenols and benzoquinones: characterisation and biosynthetic pathway. Molecules. 2013;18:11429–11451. doi: 10.3390/molecules180911429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruther, Podsiadlowski & Hilker (2001).Ruther J, Podsiadlowski L, Hilker M. Quinones in cockchafers: additional function of a sex attractant as an antimicrobial agent. Chemoecology. 2001;11:225–229. doi: 10.1007/PL00001855. [DOI] [Google Scholar]
- Ruther et al. (2001).Ruther J, Reinecke A, Tolasch T, Hilker M. Make love not war: a common arthropod defence compound as sex pheromone in the forest cockchafer Melolontha hippocastani. Oecologia. 2001;128:44–47. doi: 10.1007/s004420100634. [DOI] [PubMed] [Google Scholar]
- Sander & Hall (2015).Sander S, Hall D. Variation in opsin genes correlates with signalling ecology in North American fireflies. Molecular Ecology. 2015;24:4679–4696. doi: 10.1111/mec.13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliger et al. (1961).Seliger H, McElroy W, White E, Field G. Stereo specificity and firefly bioluminescence, a comparison of natural and synthetic luciferins. Proceedings of the National Academy of Sciences of the United States of America. 1961;47:1129–1134. doi: 10.1073/pnas.47.8.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe et al. (2015).Sharpe ML, Dearden PK, Gimenez G, Krause KL. Comparative RNA seq analysis of the New Zealand glowworm Arachnocampa luminosa reveals bioluminescence-related genes. BMC Genomics. 2015;16:825. doi: 10.1186/s12864-015-2006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shear (2015).Shear WA. The chemical defenses of millipedes (diplopoda): biochemistry, physiology and ecology. Biochemical Systematics and Ecology. 2015;61:78–117. doi: 10.1016/j.bse.2015.04.033. [DOI] [Google Scholar]
- Smirnova, Samsonova & Ugarova (2016).Smirnova DV, Samsonova JV, Ugarova NN. The bioluminescence resonance energy transfer from firefly luciferase to a synthetic dye and its application for the rapid homogeneous immunoassay of progesterone. Photochemistry and Photobiology. 2016;92:158–165. doi: 10.1111/php.12556. [DOI] [PubMed] [Google Scholar]
- Stanger-Hall, Lloyd & Hillis (2007).Stanger-Hall KF, Lloyd JE, Hillis DM. Phylogeny of North American fireflies (Coleoptera: Lampyridae): implications for the evolution of light signals. Molecular Phylogenetics and Evolution. 2007;45:33–49. doi: 10.1016/j.ympev.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Stansbury & Moczek (2014).Stansbury MS, Moczek AP. The function of Hox and appendage–patterning genes in the development of an evolutionary novelty, the Photuris firefly lantern. Proceedings of the Royal Society of London B: Biological Sciences. 2014;281:20133333. doi: 10.1098/rspb.2013.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan et al. (2015).Tan Q-Q, Zhu L, Li Y, Liu W, Ma W-H, Lei CL, Wang XP. A de novo transcriptome and valid reference genes for quantitative real-time PCR in Colaphellus bowringi. PLoS ONE. 2015;10:e0118693. doi: 10.1371/journal.pone.0118693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thancharoen, Ballantyne & Branham (2007).Thancharoen A, Ballantyne LA, Branham M. Description of Luciola aquatilis sp. nov., a new aquatic firefly (Coleoptera: Lampyridae: Luciolinae) from Thailand. Zootaxa. 2007;1611:55–62. [Google Scholar]
- Trapnell et al. (2010).Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, Van Baren M, Salzberg S, Wold B, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trice, Tyler & Day (2004).Trice E, Tyler J, Day JC. Description of pleural defensive organs in three species of firefly larvae (Coleoptera, Lampyridae) Zootaxa. 2004;768:1–11. [Google Scholar]
- Tsutomu, Hiroki & Eiichi (1989).Tsutomu M, Hiroki T, Eiichi N. Cloning and sequence analysis of cDNA for luciferase of a Japanese firefly, Luciola cruciata. Gene. 1989;77:265–270. doi: 10.1016/0378-1119(89)90074-7. [DOI] [PubMed] [Google Scholar]
- Tursch et al. (1971).Tursch B, Daloze D, Dupont M, Pasteels JM, Tricot MC. A defense alkaloid in a carnivorous beetle. Experientia. 1971;27:1380–1381. doi: 10.1007/BF02154239. [DOI] [Google Scholar]
- Tursch, Daloze & Hootele (1972).Tursch B, Daloze D, Hootele C. Alkaloid of Propylaea quatuordecimpunctata L. (Coleoptera, Coccinellidae) Chimia. 1972;26:74–75. [Google Scholar]
- Tursch et al. (1972).Tursch B, Daloze D, Pasteels JM, Cravador A, Braekman JC, Hootele C, Zimmermann D. Two novel alkaloids from the American ladybug Hippodamia convergens (Coleoptera, Coccinellidae) Bulletin des Sociétés Chimiques Belges. 1972;81:649–650. [Google Scholar]
- Tyler et al. (2008).Tyler J, Mckinnon W, Lord GA, Hilton PJ. A defensive steroidal pyrone in the Glow-worm Lampyris noctiluca L. (Coleoptera: Lampyridae) Physiological Entomology. 2008;33:167–170. doi: 10.1111/j.1365-3032.2007.00610.x. [DOI] [Google Scholar]
- Unruh, Xu & Kramer (1998).Unruh LM, Xu R, Kramer KJ. Benzoquinone levels as a function of age and gender of the red flour beetle, Tribolium castaneum. Insect Biochemistry and Molecular Biology. 1998;28:969–977. doi: 10.1016/S0965-1748(98)00085-X. [DOI] [Google Scholar]
- Villaverde, Juárez & Mijailovsky (2007).Villaverde ML, Juárez MP, Mijailovsky S. Detection of Tribolium castaneum (Herbst) volatile defensive secretions by solid phase microextraction–capillary gas chromatography (SPME-CGC) Journal of Stored Products Research. 2007;43:540–545. doi: 10.1016/j.jspr.2007.03.003. [DOI] [Google Scholar]
- Viviani et al. (2004).Viviani V, Arnoldi FG, Brochetto-Braga M, Ohmiya Y. Cloning and characterization of the cDNA for the Brazilian Cratomorphus distinctus larval firefly luciferase: similarities with European Lampyris noctiluca and Asiatic Pyrocoelia luciferases. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2004;139:151–156. doi: 10.1016/j.cbpc.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Viviani, Carmargo & Amaral (2013).Viviani VR, Carmargo IA, Amaral DT. A transcriptional survey of the cDNA library of Macrolampis sp2 firefly lanterns (Coleoptera: Lampyridae) Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. 2013;8:82–85. doi: 10.1016/j.cbd.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Wang et al. (2007).Wang Y, Fu X, Lei C, Jeng M-L, Nobuyoshi O. Biological Characteristics of the Terrestrial Firefly Pyrocoelia pectoralis (Cleoptera: Lampyridae) The Coleopterists Bulletin. 2007;61:85–93. doi: 10.1649/907.1. [DOI] [Google Scholar]
- White, McCapra & Field (1963).White EH, McCapra F, Field GF. The structure and synthesis of firefly luciferin. Journal of the American Chemical Society. 1963;85:337–343. doi: 10.1021/ja00886a019. [DOI] [Google Scholar]
- Zhang et al. (2015).Zhang J, Wang B, Dong S, Cao D, Dong J, Walker WB, Liu Y, Wang G. Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta. PLoS ONE. 2015;10:e0117054. doi: 10.1371/journal.pone.0117054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Zhao & Yang (2012).Zhu J-Y, Zhao N, Yang B. Global transcriptome profiling of the pine shoot beetle, Tomicus yunnanensis (Coleoptera: Scolytinae) PLoS ONE. 2012;7:e32291. doi: 10.1371/journal.pone.0032291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
The raw data has been supplied as Supplementary File.