Figure 1.
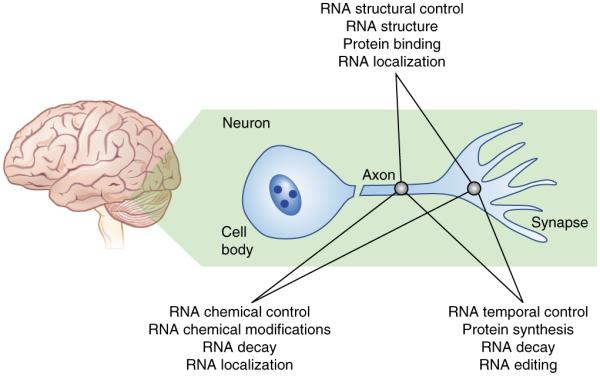
In different local environments of the neuron, epitranscriptomic mechanisms can be employed independently and bidirectionally to regulate the qualitative state of RNA and effect experience-dependent changes in neuronal function in the brain. The intersection of various aspects of RNA control, within differing regions of the same cell, affords both mechanistic and spatial control over RNA metabolism. RNA trafficking within axons to the synapse is controlled by RNA-binding proteins, which recognize unique structural and sequence elements in RNAs. Protein synthesis is controlled temporally to guide protein abundance and synapse formation. Checkpoint mechanisms may be brought about by RNA modifications, which have already been demonstrated to regulate RNA decay. Each of these are interconnected and therefore substantially increase the complexity of RNA regulation in neurons.
