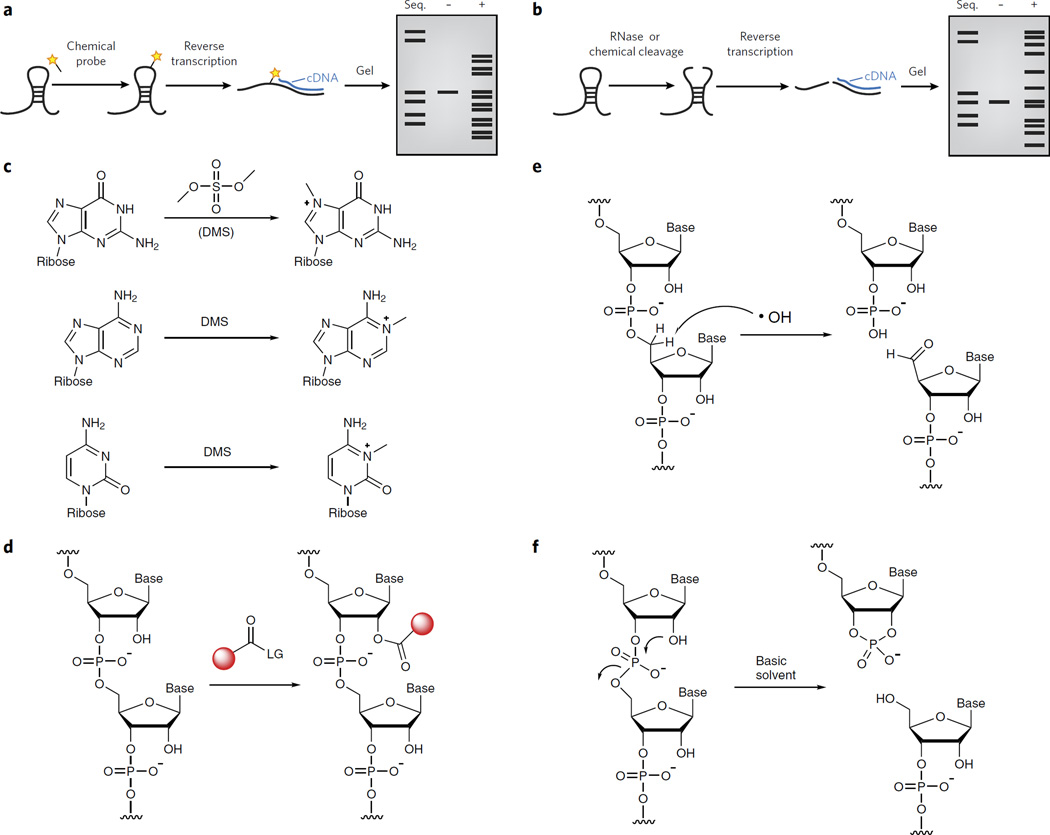
Figure 2. Chemical methods to probe RNA structure.
(a) A depiction of a typical chemical probing experiment. The RNA is first folded and then treated with a reagent that reacts covalently. The site of reaction is read out by reverse transcription and synthesis of cDNA that can be resolved by denaturing gel electrophoresis. (b) A depiction of a typical RNA structure probing experiment in which RNA cleavage is used to probe structure. The RNA is first folded and then treated with a reagent that cleaves the RNA backbone. The site of cleavage is read out by reverse transcription and synthesis of cDNA that can be resolved by denaturing gel electrophoresis. (c) Dimethylsulfate can alkylate the N7 of guanosine, the N1 of adenosine and the N3 of cytidine. (d) RNA SHAPE analysis measures the propensity of the 2′-OH to become activated as a nucleophile and undergo an acylation reaction. (e) Hydroxyl radical probing at the C5′ position leads to strand cleavage, resulting in the formation of a 3′-phosphate and 5′-aldehyde. (f) In-line probing results in a 2′,3′-cyclic phosphate and a 5′-hydroxyl.