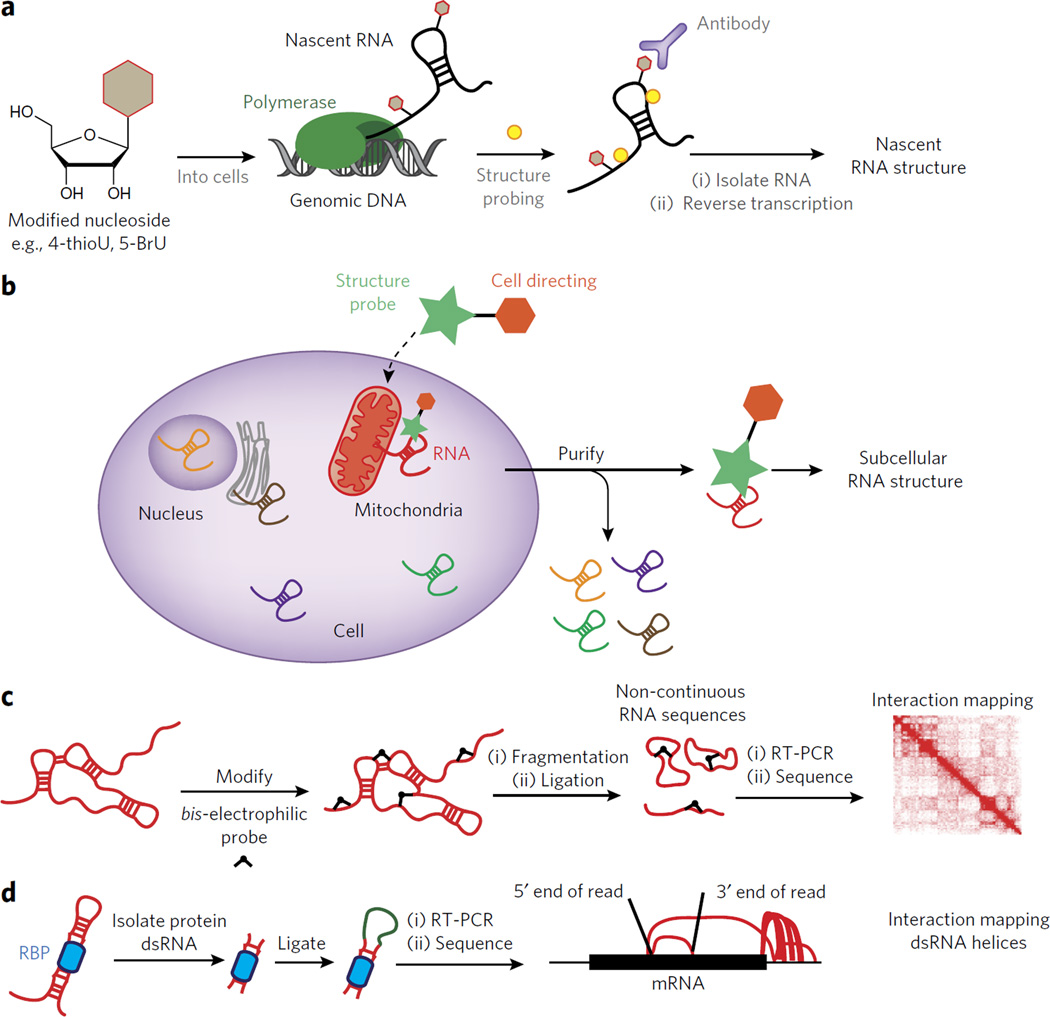
Figure 5. Outstanding challenges for understanding RNA structure inside living cells.
(a) Outline of an experiment for interrogating RNA structure formation during transcription. In such a case a modified nucleoside can be introduced into the cell and then used to enrich for co-transcriptionally probed RNA structure. (b) A schematic for the design of a dual-functioning chemical probe to measure RNA structure within unique subcellular compartments. (c) A depiction of how a chemical probe can be used to identify three-dimensional contacts within folded RNAs. The results are mapped to an interaction map, depicting the spatial relationship between two points in the RNA sequence. (d) A schematic for the recently developed method known as hiCLIP84. In hiCLIP non-contiguous reads are mapped to genes and represented by rainbow maps to connect primary sequence points through space.