FIG 2.
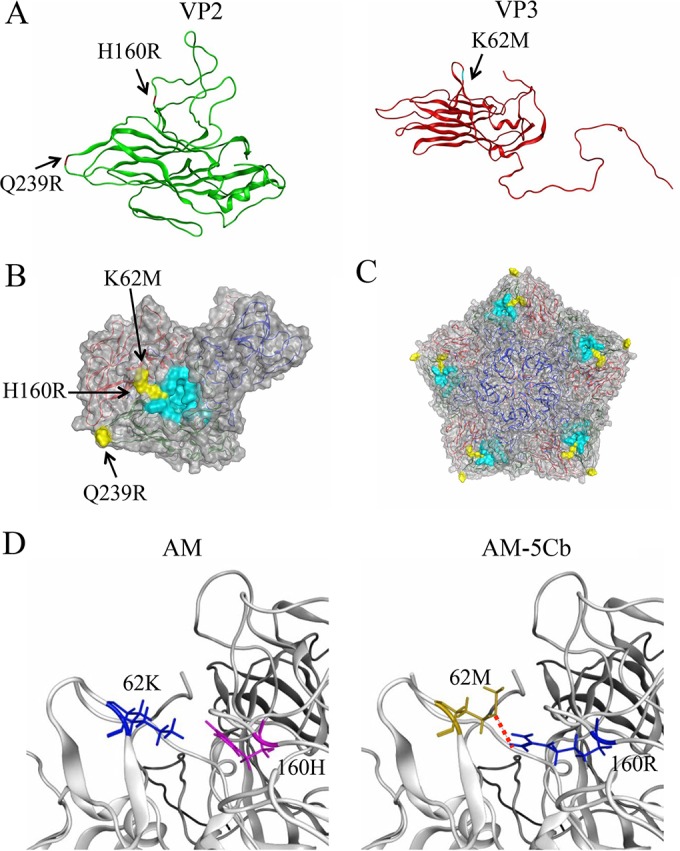
Structural models of SAFV-3 capsid proteins. The models were constructed by homology modeling based on the X-ray-derived crystal structure of the virus particle of the TMEV DA strain. (A) Models of the VP2 (green) and VP3 (red) proteins of the AM-5Cb strain. Substituted residues in the AM-5Cb strain indicated are H160R at the loop of the VP2 alias puff B structure, Q239R at the loop of VP2, and K62M at the loop in the VP3 knob structure. (B, C) Models of the VP1, VP2, and VP3 complex (B) and the pentamer of the complex (C) on the virion surface. Three substituted residues in the AM-5Cb strain (yellow) and residues of the receptor binding region of TMEV (cyan) are highlighted. (D) Closeup view around the putative receptor binding domain of the SAFV-3 VP2 and VP3 proteins: AM (left) and AM-5Cb (right) strains. A red dotted line indicates that a novel hydrogen bond forms between side chains of the mutated residues (VP2-160R and VP3-62M) in the AM-5Cb strain.
