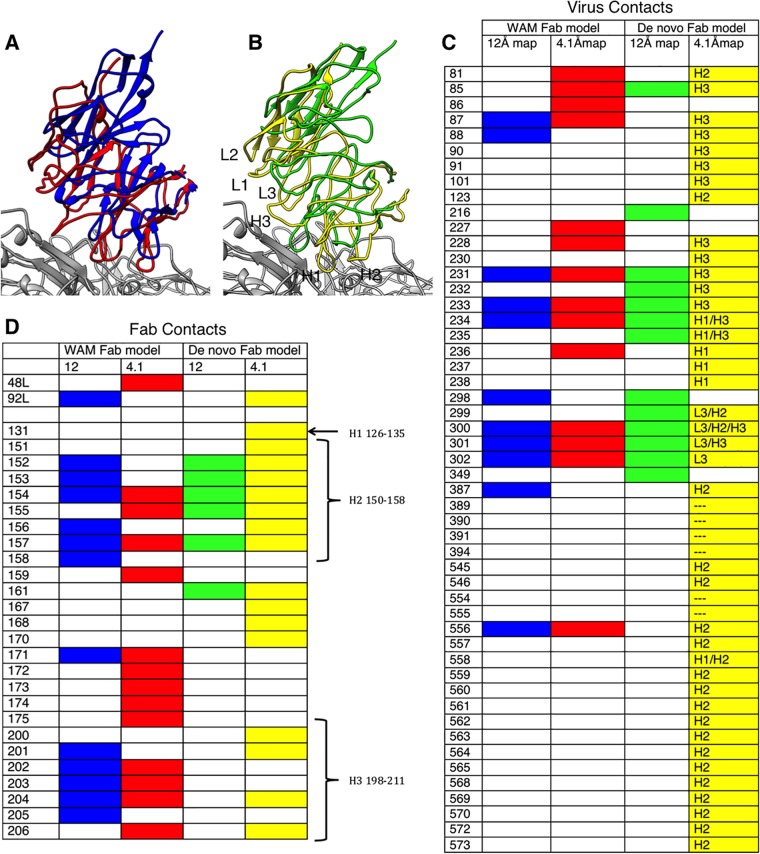
FIG 5.
Effect of resolution on virus-Fab interface identification based on the final model compared to previous models. (A) WAM E fitted into the previous low-resolution map (blue) (19) superimposed on WAM E fitted into the new high-resolution map (red). (B) The de novo Fab structure fitted into the previous low-resolution map (green) superimposed on the de novo Fab structure built into the new high-resolution map (yellow). (C, D) Contacts on the surface of the virus and Fab, respectively, are identified from the fitted structures, with colors corresponding to the ribbon color of each fitted Fab. The list of contacts on the virus surface also indicates which antibody complementarity-determining region (CDR) loop is the binding partner: L3, H1, H2, H3, or none (---). The CDR loops are also labeled on the Fab footprint.