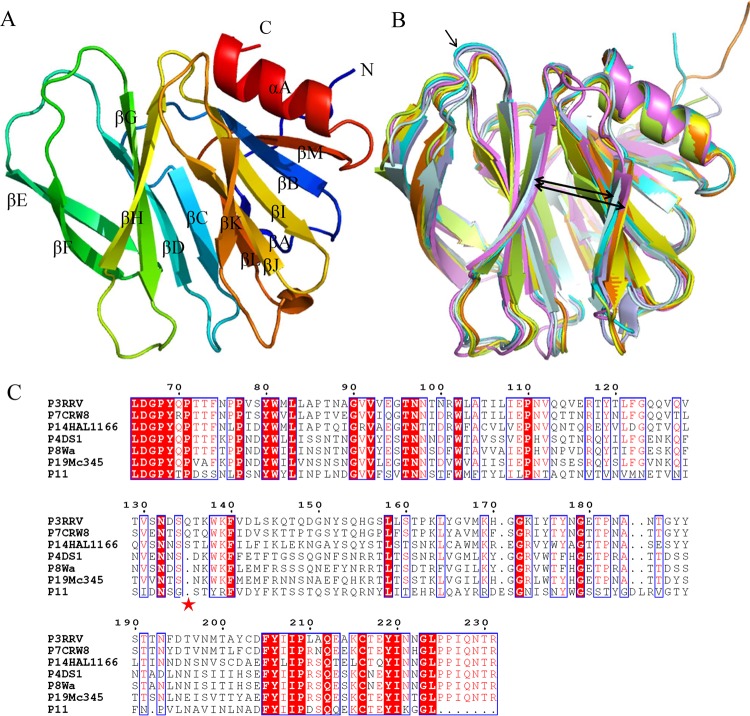
FIG 4.
Structural analysis of human P[19] Mc345 VP8*. (A) Overall structure of human P[19] VP8* with two twisted antiparallel sheets consisting of strands A, L, D, G, and H and M, B, I, J, and K. (B) Superimposition of Mc345 human P[19] VP8* structure (5GJ6) (lemon) on those of rhesus rotavirus (RRV) P[3] (1KQR) (pale cyan), HAL1166 P[14] (4DRV) (cyan), CRW-7 P[7] (2I2S) (light blue), DS-1 P[4] (2AEN) (orange), Wa P[8] (2DWR) (yellow), and human rotavirus (HRV) P[11] (4YG0) (violet). The width of the cleft that separates the two β-barrel sheets is shown by the thick arrows. Residue 135 of P[3] and P[14], which was deleted in P[4], P[8], P[11], and P[19], is indicated by the thin arrow. (C) Sequence alignment of VP8* proteins of RRV P[3], HAL1166 P[14], CRW-7 P[7], DS-1 P[4], Wa P[8], P[11], and Mc345 P[19]. The position of residue 135 is labeled with an asterisk. The alignment was done with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/), and the colors and box labels were generated with ESPript (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). Red shading indicates residues that are the same in all the aligned sequences, and red letters indicate the residues with high conservation.