FIG 3.
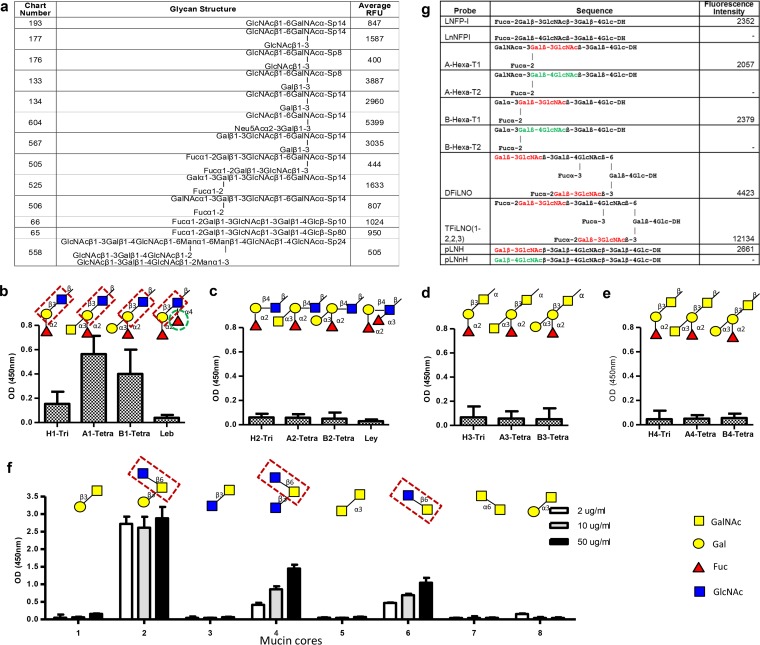
P[19] VP8* recognizes mucin cores 2, 4, and 6 and type 1 HBGAs. (a) A list of top glycans recognized by P[19] VP8* in a glycan array containing 610 glycans. (b to e) Binding to four types (1 to 4) of HBGA glycans, of which P[19] VP8* recognized only type 1 HBGA glycans. The signals for binding to the H1 trisaccharide (Fucα1-2Galβ1-3GlcNAc-) were weak to moderate but significantly increased with the addition of the A or B residues, whereas further addition of a β1-3 fucose (the Lewis epitope, green dashed circle) to the GlcNAc (forming Leb) completely blocked the binding. The disaccharide Galβ1-3GlcNAc of the type 1 HBGA precursor (highlighted in red dashed boxes) is believed to be essential for binding, while the terminal fucose, A, or B residues added to Gal [Fucα1-2Galβ1-3GlcNAc-, GalNAcα1-3(Fucα1-2)Galβ1-3GlcNAc-, and Galα1-3(Fucα1-2)Galβ1-3GlcNAc-] could increase the binding. (f) Among the eight mucin core glycans (core 1 to core 8) tested, only mucin cores 2, 4, and 6 revealed binding signals. The disaccharide GlcNAcβ1-6GalNAc is believed to be essential for binding, which is highlighted in red dashed boxes. The ELISA was performed in triplicate, and the whole experiment was repeated once. The error bars represent the standard deviations from triplicate wells. (g) Analysis with a focused array of lipid-linked sequence-defined probes, which was carried out to further determine the HBGA binding specificity of P[19] VP8*. Six pairwise comparisons of type 1 and type 2 chains with different lengths and capping moieties are shown. P[19] VP8* recognized only the type 1-containing probes (red). The type 2 chain probes are marked green. DH represents the amino lipid that the oligosaccharide probes were linked to. A dash indicates fluorescence intensity less than 1.