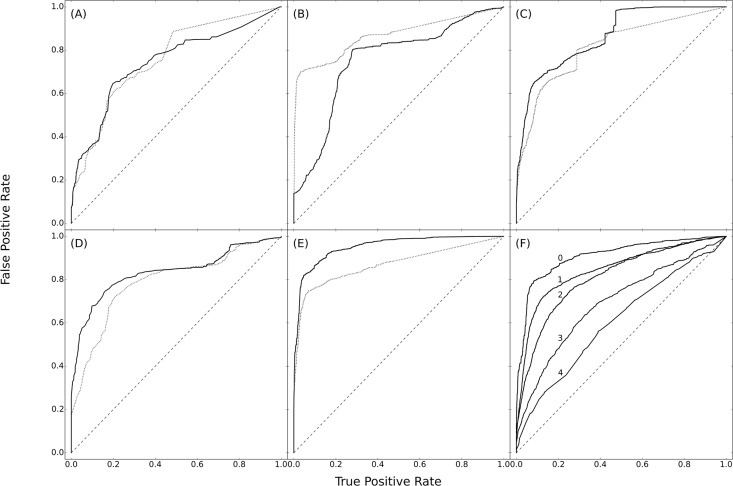
Fig 5. Classification performance of similarity functions.
Pairwise similarities were calculated, using the indicated similarity functions, for all RNAs in the curated dataset and ranked from high to low. A pair of RNAs from the same curated family is considered a positive match; otherwise they are considered to be a negative match. In all panels, the dashed line indicates the simple fingerprint, and the solid line the extended fingerprint. The AUC for the simple and extended fingerprints, respectively, are indicated in parentheses, below. (A) Intersection Similarity (AUC simple, 0.759; extended, 0.746), (B) Cosine Similarity (0.867; 0.753), (C) Dice Similarity (0.821; 0.864), (D) Hamming Similarity (0.789; 0.834), and (E) Jaccard Similarity (0.870; 0.952). (F) Classification after random removal of vertices from RNA graphs. All RNAs (except for tRNA and 5S rRNA which are too small for 70% stem removal) are included. The five lines show ROC curves with differing fractions of stems removed (AUC in parentheses): (0) no stem removal (AUC = 0.909), (1) 10% stem removal (0.844), (2) 30% stem removal (0.810), (3) 50% stem removal (0.691), and (4) 70% stem removal (0.605).