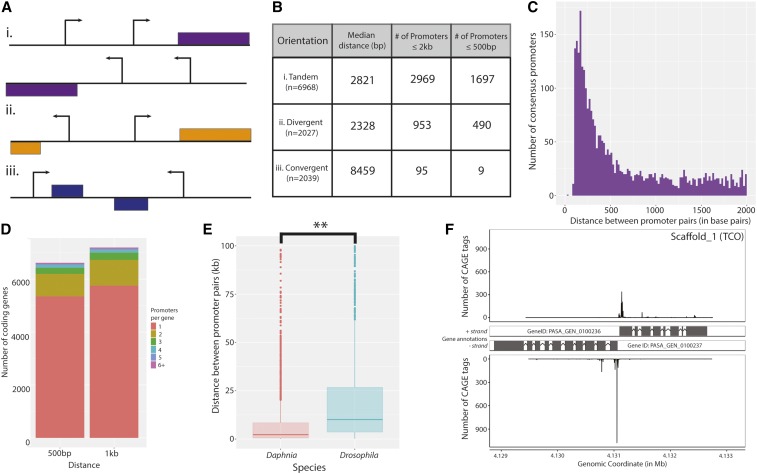
Figure 3.
Compact promoter orientations in D. pulex. (A) Illustration of possible configurations of promoter pairs. Promoters can be found in (i) tandem, (ii) divergent, and (iii) convergent orientations. (B) Distance measures between promoter pairs for each of the orientations in (A) are shown. The median distance (in bp) of promoter pairs in (i) tandem, (ii) divergent, and (iii) convergent orientations are listed, in addition to the number of consensus promoter pairs within 2 kb and 500 bp. (C) A histogram of the distribution of distances between tandem promoter pairs is shown, which reveals a large number of closely-spaced consensus promoters. Only those promoter pairs within 2 kb are shown. (D) Putative alternative promoter usage in D. pulex. Two barplots representing annotated coding genes by number of associated consensus promoters (left, within 500 bp; right, within 1 kb of the annotated translation start site) are shown. (E) Divergent promoters in D. pulex are more closely-spaced than those in D. melanogaster. Boxplots representing the overall distances between convergent promoters in D. pulex (coral) and D. melanogaster (cyan) are shown. (** P < 0.001 Welch two sample t-test) (F) A representative example of closely-spaced divergent promoter pairs in D. pulex. Aligned CAGE reads from the selected coordinates (4,129,000–4,133,122 on scaffold 1) are shown, with the CAGE tag abundance (in number of CAGE tags) presented on the y-axis. CAGE, Cap Analysis of Gene Expression.