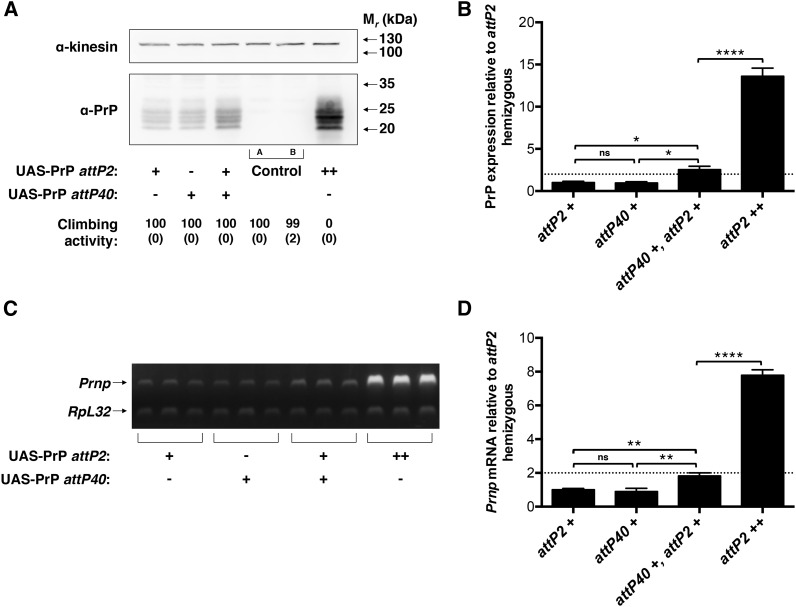
Figure 2.
Allelic interaction between UAS-PrP transgenes is required for enhanced Prnp transcriptional efficacy. UAS-PrP inserted at attP2 and/or attP40 was expressed using the Cha-GAL4 driver. (A) Western blot of fly head homogenates (+, hemizygous; ++, homozygous at the indicated locus). Note that PrP is independently expressed at approximately equal levels from attP2 and attP40. Experiment was performed in triplicate with a representative blot shown. Climbing activity of each line is indicated below the corresponding lane of the Western blot and expressed as percent passing the climbing assay (see Materials and Methods) with standard deviation shown below in brackets. Control A is driver alone; control B is UAS-PrP alone. (B) Densitometry of Western blot signals in (A) and associated replicates. PrP signals were normalized to the kinesin loading control and then to the hemizygous attP2 UAS-PrP line. (C) Semiquantitative RT-PCR performed on fly head mRNA of attP2 and attP40 UAS-PrP constructs driven by Cha-GAL4 (+, hemizygous; ++, homozygous at the indicated locus). Prnp and RpL32 transcripts were coamplified in the same PCR reactions, and each lane represents a biological replicate. (D) Densitometry of RT-PCR signals in (C). For each sample, the ratio of Prnp to RpL32 mRNA levels was taken and then normalized to the average value for the hemizygous attP2 UAS-PrP line. Error bars in (B) and (D) represent standard deviations, with n = 3 for each group; ns denotes note significant, ** P < 0.01, and **** P < 0.0001. Dotted line emphasizes a twofold increase in expression relative to hemizygous. mRNA, messenger RNA; PrP, prion protein; RT-PCR, reverse transcription polymerase chain reaction.