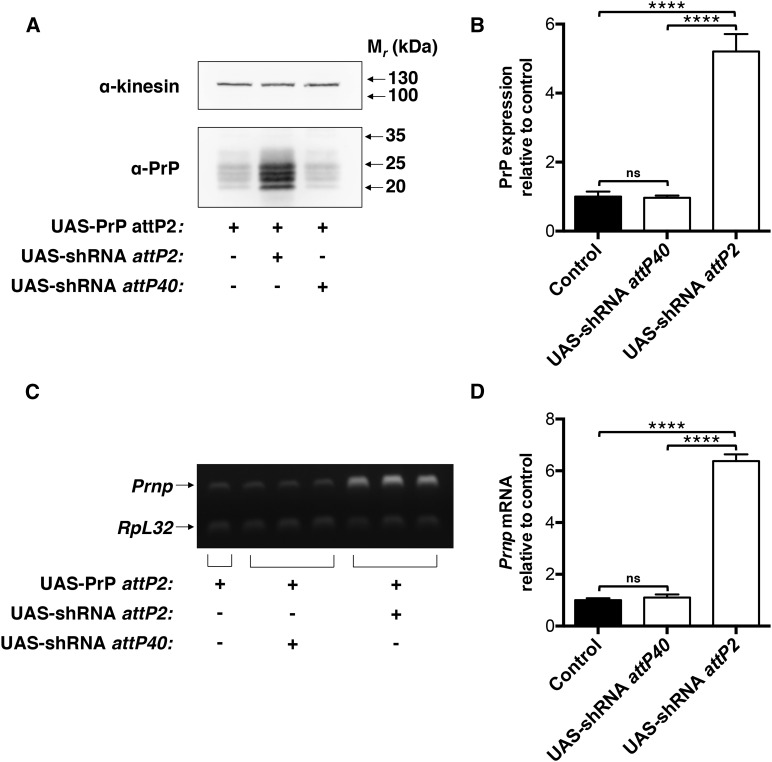
Figure 4.
Control UAS-shRNA constructs from the Drosophila TRiP library enhance UAS-PrP transcriptional efficacy in trans. In flies expressing UAS-PrP from attP2 driven by Cha-GAL4, an shRNA hairpin against GFP was expressed at the paired attP2 or at the unpaired attP40 locus to assess the suitability of the TRiP library as an in vivo screening tool for transvection activity. (A) Western blot of fly head homogenates. Experiment was performed in triplicate with a representative blot shown. (B) Densitometry of Western blot signals in (A) and associated replicates. PrP signals were normalized to the kinesin loading control and then to the hemizygous attP2 UAS-PrP line lacking UAS-shRNA. (C) Semiquantitative RT-PCR performed on fly head mRNA. Prnp and RpL32 transcripts were coamplified in the same PCR reactions, and each lane represents a biological replicate. (D) Densitometry of RT-PCR signals in (C). For each sample, the ratio of Prnp to RpL32 mRNA levels was taken and then normalized to the average value for the attP2 UAS-PrP line lacking UAS-shRNA. Error bars in (B) and (D) represent standard deviations (n = 2 for UAS-PrP alone mRNA quantification, n = 3 for all other groups); ns denotes not significant and **** P < 0.0001. GFP, green fluorescent protein; mRNA, messenger RNA; PrP, prion protein; RT-PCR, reverse transcription polymerase chain reaction; shRNA, short hairpin RNA; TRiP, Transgenic RNA Interference Project.