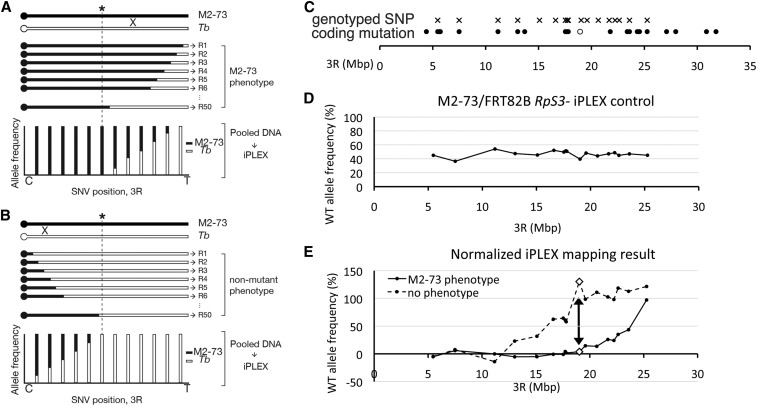
Figure 4.
iPLEX MassARRAY genotyping maps the M2-73 mutant. (A, B) Recombinant mapping principles. ‘X’ illustrates that meiotic recombination occurs between homologs. (A) A cartoon representation of recombinant chromosome arms 3R expected to retain the M2-73 phenotype. These should retain M2-73 SNPs proximal to and including the locus responsible (asterisk), and exhibit various crossover points distal to the locus. When DNA from 50 such recombinants is pooled, the frequency of M2-73 SNPs is expected to decrease distal to the locus as the frequency of Tb SNPs increases. (B) A cartoon representation of recombinant chromosome arms 3R expected to lack the M2-73 phenotype. These should retain Tb SNPs distal to and including locus responsible for the M2-73 phenotype, and exhibit crossover points proximal to the locus. When DNA from such recombinants is pooled, the frequency of M2-73 SNPs should increase proximally to the locus as the frequency of Tb SNPs decreases. Comparing observed SNP frequencies for the two pools should limit the mutant locus to a chromosome segment that retains 100% M2-73 SNPs in phenotypically mutant recombinants and 100% Tb SNPs in phenotypically wild-type recombinants. (C) Distribution of the 17 SNPs that were assessed by iPLEX (crosses) and 22 exonic variants on the M2-73 chromosome (circles). An open circle shows the position of the coding mutation in Xrp1, closed circles show the other mutations. Some of the coding alleles were used as SNPs, in addition to noncoding SNPs. (D) Allele frequencies determined by iPLEX for 17 SNPs in the FRT82B M2-73/FRT82B P{w+, arm-LacZ} P{w+, tub-Gal80} RpS3 genotype. Deviation from 50% reflects amplification and detection bias in the iPLEX method (see section on Mapping a single allele using recombination and iPLEX MassARRAY). This heterozygote genotype is used because the recombinant DNA is also heterozygous with the FRT82B P{w+, arm-LacZ} P{w+, tub-Gal80} RpS3 chromosome, in which background the M2-73 phenotype is assessed. (E) Normalized frequencies of reference alleles (derived from the Tb1 chromosome). Solid line, pooled recombinants exhibiting the M2-73 phenotype; dashed line, pooled recombinants lacking the M2-73 phenotype. Normalized allele frequency is calculated as (measured % Tb allele in recombinants)/(measured % Tb allele in (D) control) × 100, to account for any amplification or detection bias in the MassARRAY method. The arrow indicates SNP 19,029,906 (+BDGP Release 6) where Tb SNPs are recovered in ∼0% of phenotypically M2-73 recombinants and ∼100% of phenotypically wild-type recombinants. This SNP is ∼100 kb from a coding mutation in the Xrp1 gene (open circle in A).