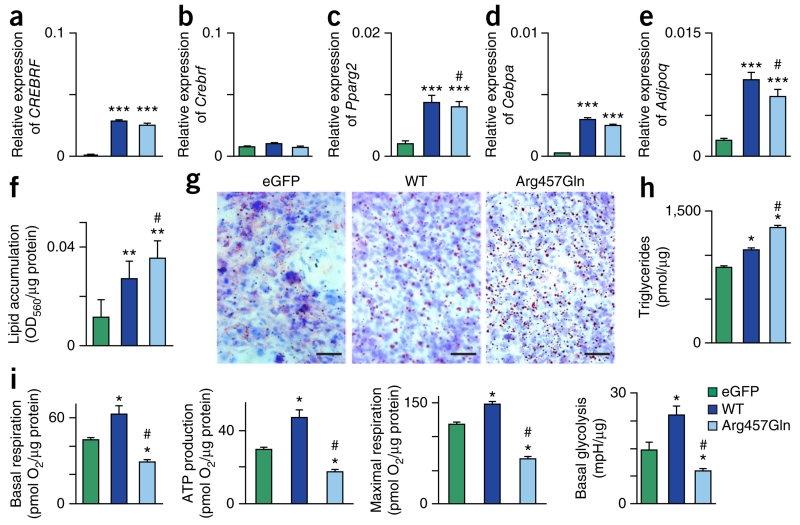
Figure 2.
CREBRF variants, adipogenic differentiation, lipid accumulation, and energy homeostasis. 3T3-L1 mouse preadipocytes overexpressing enhanced GFP–only negative control (eGFP), wild-type human CREBRF (WT), or Arg457Gln human CREBRF were collected at 8 d after confluence in the absence of hormonal stimulation of adipogenic differentiation. (a–e) mRNA levels of human CREBRF (a) and endogenous mouse Crebrf (b), Pparg2 (c), Cebpa (d), and Adipoq (e) relative to those of the β-actin (Actb) reference gene determined using quantitative RT–PCR. Values are given as means ± s.e.m. from three biological replicates with four technical replicates each (n = 3 × 4 = 12). Representative results from one of four experiments are shown. (f) Quantification of lipid accumulation with Oil Red O staining normalized to protein content (OD560/μg protein). Data are shown as means ± s.e.m. from three transfection replicates with eight wells for each transfection (n = 3 × 8 = 24). (g) Representative photomicrographs of Oil Red O staining to visualize lipid droplets (red) with counterstaining of nuclei with hematoxylin (blue). Scale bars, 50 μm. (h) Biochemical assay for triglycerides. Data are shown as means ± s.e.m., n = 2 biological replicates. (i) Key bioenergenic variables as determined on the basis of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) normalized to protein content. Values are given as means ± s.e.m. (n = 6 biological replicates). mpH, 0.01 pH unit. Statistical analysis: one-way analysis of variance (ANOVA), two-sided Games–Howell post-hoc test. *P < 0.03, **P < 1 × 10−3, ***P < 1 × 10−4 compared to 3T3-L1 cells transfected with eGFP control construct; #P < 0.05 compared to 3T3-L1 cells transfected with construct for wild-type CREBRF.