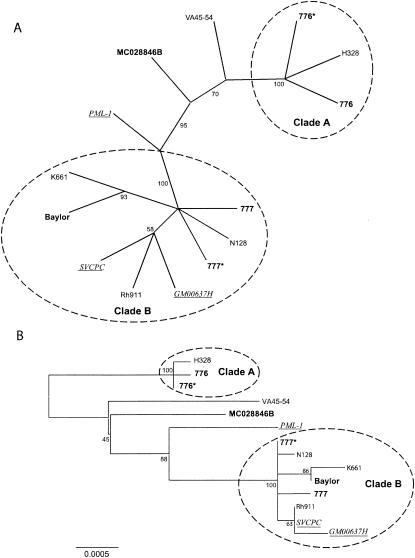
FIG. 1.
Phylogenetic tree for SV40 based on complete genome sequences. (A) Unrooted consensus tree of 1,000 bootstrap replicates of the whole-genome data, generated by the maximum-parsimony method. Conventions for labels are as follows: monkey isolates are in roman type; vaccine isolates are in boldface type; human isolates are in underlined italic type. (B) Consensus tree of 1,000 bootstrap replicates of the whole-genome data, generated by the neighbor-joining method. Distances are proportional to the number of mutational changes, after the Kimura (20) correction has been applied; the scale is in proportion of changes relative to the whole genome. The scale bar (0.0005) represents the number of substitutions per site. Numbers proximal to nodes in the tree indicate the proportion of 1,000 bootstrap replications; only bootstrap values >50% are shown. Two clades (A and B) are resolved, with several outlier sequences.