Figure 1.
Single-Cell Transcriptome Analyses of Human Pancreas
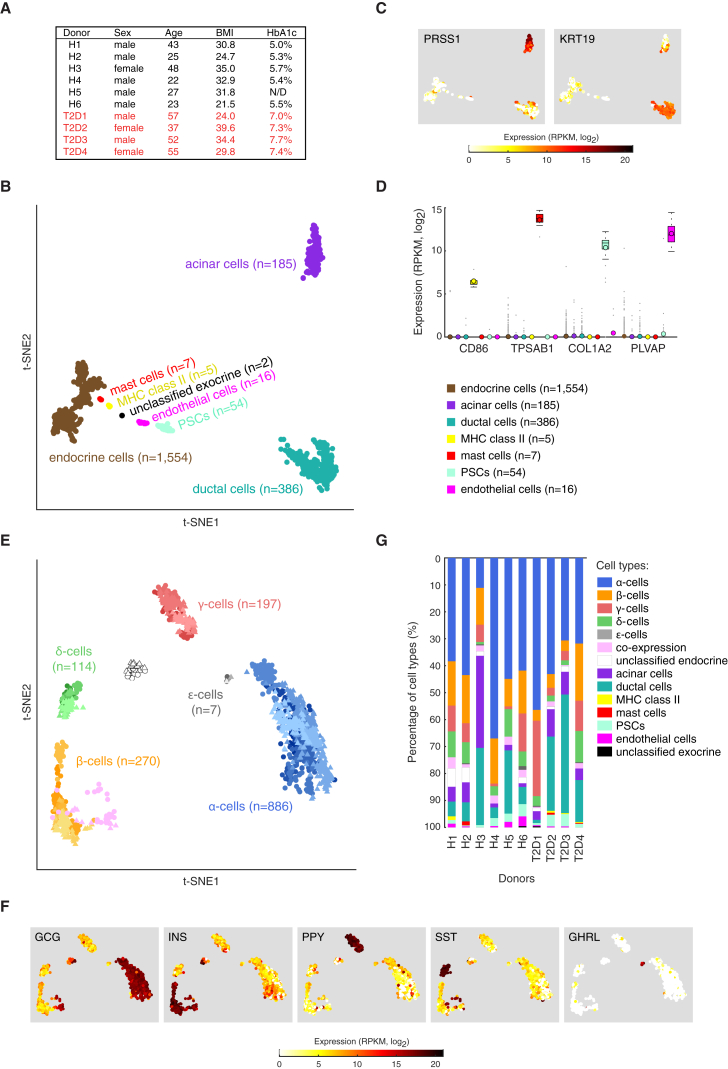
(A) Table of donor information (HbA1c, glycated hemoglobin).
(B) Projection of all cells (n = 2,209) onto two dimensions using t-SNE based on the expression values (log2RPKM) of the 1,000 genes with highest biological variation across cells.
(C) Expression (log2RPKM) of exocrine marker genes (PRSS1 for acinar and KRT19 for ductal cells) overlaid onto the 2D t-SNE as shown in (B).
(D) Boxplots displaying the expression levels in the seven obtained clusters of marker genes for MHC class II antigen-presenting cells (CD86), mast cells (TPSAB1), pancreatic stellate cells (COL1A2), and endothelial cells (PLVAP). Median and mean are shown as a line and circle, respectively. Edges of each box indicate the 25th and 75th percentiles. Bars extend to extreme data points and outliers are plotted as gray dots.
(E) Two-dimensional t-SNE projection of the endocrine cells (n = 1,554) based on the expression values (log2RPKM) of the 500 genes with highest biological variation across endocrine cells. The obtained clusters were assigned to the endocrine cell types based on the hormone expression levels in (F). Colors correspond to cell types and shadings indicate donors. Healthy and T2D cells are marked with circles and triangles, respectively.
(F) t-SNE representation of cells as in (E) illustrating the expression of the five endocrine hormones: GCG, INS, PPY, SST, and GHRL. Color scale is according to log2RPKM values, with white and red colors corresponding to minimum (zero) and maximum (log2RPKM = 21) expression, respectively.
(G) Bar graphs showing the percentage of cells classified into cell types, per donor.