Figure 3. TUT‐DIS3L2 pathway targets precursor and aberrant forms of tRNAs.
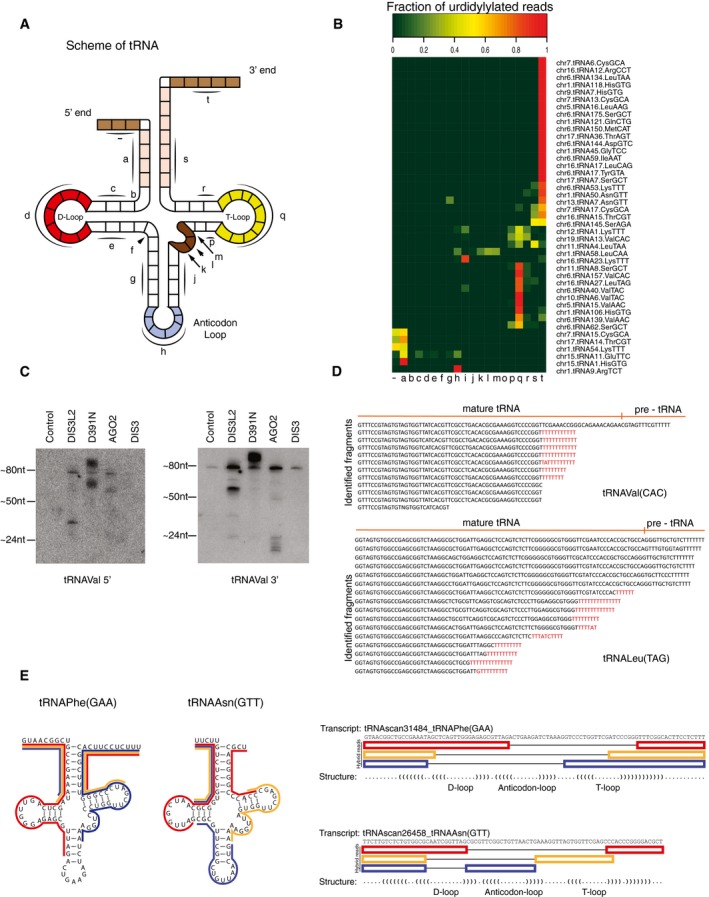
- Schematic representation of the pre‐tRNA secondary structure.
- Uridylated tRNA reads overlap mainly with 3′ trailers, T‐loop, and anticodon loop regions. Heat map showing the coverage of uridylated reads on pre‐tRNAs. The pre‐tRNA subregions (as indicated in A) are in small letters on the x‐axis.
- Northern blot analysis of RNAs immunoprecipitated with Flag‐tagged WT (DIS3L2) and D391N (MUT) DIS3L2, AGO2, and DIS3, respectively. The blots were probed with radioactively labeled probes for 5′ and 3′ ends of mature tRNAVal, respectively.
- Sequencing of MUT DIS3L2‐bound tRNAVal and tRNALeu confirmed the uridylation status of the tRNA fragments identified by CLIP‐seq. Genomic sequence for each tRNA is indicated at the top with the 3′ trailer region separated by a vertical line. Non‐templated uridine residues are in red.
- Identification of hybrid reads (produced by intrastrand ligation of RNaseT1 products during CLIP protocol) mapping to tRNAPhe(GAA) and tRNAAsn(GTT) genes. To the right, the schematic representation of selected hybrid reads with corresponding color‐code.
