Summary
Head blight caused by Fusarium graminearum threatens world‐wide wheat production, resulting in both yield loss and mycotoxin contamination.
We reconstructed the global F. graminearum gene regulatory network (GRN) from a large collection of transcriptomic data using Bayesian network inference, a machine‐learning algorithm. This GRN reveals connectivity between key regulators and their target genes. Focusing on key regulators, this network contains eight distinct but interwoven modules. Enriched for unique functions, such as cell cycle, DNA replication, transcription, translation and stress responses, each module exhibits distinct expression profiles.
Evolutionarily, the F. graminearum genome can be divided into core regions shared with closely related species and variable regions harboring genes that are unique to F. graminearum and perform species‐specific functions. Interestingly, the inferred top regulators regulate genes that are significantly enriched from the same genomic regions (P < 0.05), revealing a compartmentalized network structure that may reflect network rewiring related to specific adaptation of this plant pathogen.
This first‐ever reconstructed filamentous fungal GRN primes our understanding of pathogenicity at the systems biology level and provides enticing prospects for novel disease control strategies involving the targeting of master regulators in pathogens. The program can be used to construct GRNs of other plant pathogens.
Keywords: Bayesian network inference, cell circuits, Fusarium graminearum, modularity, network rewire and fungal pathogenesis
Introduction
Fusarium head blight (FHB), caused by the filamentous ascomycete Fusarium graminearum (F. graminearum), is one of the most devastating diseases in wheat (Triticum aestivum), barley (Hordeum vulgare) and other small grains around the world (Goswami & Kistler, 2004; Leslie & Summerell, 2006; Rep & Kistler, 2010; Guo & Ma, 2014). FHB results in yield loss and contamination of grains with mycotoxins (Leslie & Summerell, 2006), fungal secondary metabolites toxic to animals, including humans. The management of FHB remains challenging because of a lack of effective resistant wheat cultivars and the prevalence of pathogen resistance to fungicides. The understanding of F. graminearum pathobiology at the systems level is vital to effective disease and mycotoxin management.
From a systems biology perspective, any biological process, such as growth, reproduction or host invasion, is accomplished by genome‐encoded molecular components that are orchestrated and assembled into interconnected cell circuits (Kim et al., 2009). A gene regulatory network (GRN) depicts the relationships between regulatory components (e.g. kinases and transcription factors (TFs)) and their target genes (e.g. enzymes and structural proteins), key components of cell circuits. Among fungal species, GRN studies have extensively focused on yeasts, such as Saccharomyces cerevisiae (Pe'er et al., 2001, 2002; Guelzim et al., 2002; Lee et al., 2002; Segal et al., 2003; Nachman et al., 2004; Kim et al., 2006; Hu et al., 2007; Darabos et al., 2011) and Candida albicans (Homann et al., 2009; Ramachandra et al., 2014). By contrast, the reconstruction of GRN for filamentous fungi remains in its infancy. The best understood F. graminearum transcriptional regulation system is the Tri gene cluster, which is modulated by two internal TFs, Tri6 and Tri10 (Seong et al., 2009; Nasmith et al., 2011), and directs the biosynthesis of trichothecene mycotoxins (Goswami & Kistler, 2004; Rep & Kistler, 2010). A chromatin immunoprecipitation sequencing (ChIP‐seq) analysis (Nasmith et al., 2011) revealed that Tri6 physically binds to the promoter sequences of six Tri genes and 192 non‐Tri genes by recognizing the ‘TNAGGCC’ motif (Nasmith et al., 2011). It remains to be determined how cellular components work systemically to regulate F. graminearum development, invasive growth and virulence.
There are rich resources to facilitate such an endeavor. The F. graminearum genome has been fully sequenced (Cuomo et al., 2007) and putative regulatory proteins have been comprehensively annotated (Guldener et al., 2006a; Park et al., 2008, 2011; Wong et al., 2011). In addition to the Tri cluster, over 100 pathogenicity‐related genes have been characterized, including signal proteins (SPs), such as G proteins (Yu et al., 2008), small GTPases (Bluhm et al., 2007) and mitogen‐activated protein kinases (MAPKs) (Urban et al., 2003; Jenczmionka & Schafer, 2005). Genome‐wide functional studies of F. graminearum TFs (Son et al., 2011) and protein kinases (PKs) (Li et al., 2011) have linked 170 TFs and 64 PKs to fungal growth, development and virulence. An F. graminearum protein–protein interactome database (FPPI) has been constructed (Zhao et al., 2009). Furthermore, over 100 transcriptome datasets, covering diverse biological states, such as plant infection (Boddu et al., 2006; Guldener et al., 2006b; Lysoe et al., 2011), sexual development (Qi et al., 2006; Hallen et al., 2007; Lee et al., 2010; Sikhakolli et al., 2012), conidial germination (Seong et al., 2008) and mycotoxin production (Gardiner et al., 2009; Seong et al., 2009; Jonkers et al., 2012), are available in the PLEXdb public database (Dash et al., 2012) (http://www.plexdb.org). Large collections of expression profiles, either generated in our research or obtained from PLEXdb, established a foundation for the systemic inference of the F. graminearum GRN.
Many sophisticated computational programs, including Gaussian graphical models (Kishino & Waddell, 2000; Schafer & Strimmer, 2005a,b), probabilistic Boolean networks (Kauffman, 1969; Glass & Kauffman, 1973; Shmulevich et al., 2002) and Bayesian networks (BNs) (Friedman et al., 2000; Pe'er et al., 2002; Segal et al., 2003), have been developed for GRN reconstruction. This study adopted MinReg (Pe'er et al., 2002), a scalable BN model that defines regulatory relationships using expression data, and has been successfully used to reconstruct yeast (Pe'er et al., 2002) and mammalian (Amit et al., 2009) GRNs. A probabilistic graphical model, BN combines multivariate probability distributions and captures the properties of conditional independence between variables. Because of its ability to describe complex stochastic processes and to provide clear methodologies for learning from noisy observations, BN has emerged as a common and attractive model for the reconstruction of GRNs from expression data (Friedman et al., 2000; Vignes et al., 2011).
Here, we report the first genome‐wide F. graminearum GRN by harnessing available genomic, transcriptomic and functional data. Remarkably, the predicted regulators were functionally correlated with target genes. Topologically, the GRN was divided into eight distinct functional modules that were differentially regulated under various biological conditions, in agreement with the finding that each module performed distinct functions. Finally, we provide evidence that the F. graminearum regulatory network is compartmentalized in the core vs species‐specific genomic regions, providing insight into the circuit rewiring that may contribute to fungal genome evolution and specialization. Key components of the GRN are promising candidates for devising novel strategies to control this destructive disease. The computational program used here is also suitable for network inference in a broad spectrum of phytopathogenic fungi.
Materials and Methods
Strains
Fusarium graminearum Schwabe PH1 wild‐type and mutants (Supporting Information Table S1) were kindly provided by Dr Jin‐Rong Xu at Purdue University, and have been described previously (Wang et al., 2011).
RNA preparation, chip hybridization and analysis
For the 27 sets of microarray data generated in this study (Table S1), total RNAs were isolated from fungal hyphae harvested from 36‐h‐old complete medium cultures with TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer's recommendations. Microarray hybridization was performed using the Affymetrix Fungal Multigenome ExonChip, and data processing and analysis were conducted as described previously (Guo et al., 2016). Three biological replicates were conducted for each experiment. For data quality control, Pearson correlation coefficients (r) were calculated, and scatterplots were created for each possible pair of biological replicates (Fig. S1) using the R package ggally.
Combined data and normalization
One‐hundred and sixty‐six F. graminearum microarray samples, subjected to 55 experimental conditions (Table S1), were obtained from PLEXdb. As these datasets were generated from different sources, expression levels in all samples were calculated uniformly from raw microarray data, as described previously (Guo et al., 2016). The expression of each gene under each experimental condition was classified into three discrete statuses: normal, upregulated and downregulated (Table S2). The normalized data were used as input in our network inference program.
MinReg algorithm implementation
The MinReg algorithm, based on the BN model (Pe'er et al., 2006), was adapted for our GRN inference. BN learning includes two aspects: learning the parameters of the conditional probability distribution for genes as nodes and learning the BN structure. With thousands of gene nodes in a regulatory network, it is an NP (nondeterministic polynomial time)‐complete problem to enumerate all possible network structures and optimize the parameters. Under the assumption that only a certain number of regulators regulate a target gene, the MinReg algorithm uses a heuristic greedy strategy to add regulators, one by one, by selecting the regulator that can increase the BN score (BNS) the most. If adding a new regulator does not significantly increase the overall BNS, the algorithm terminates. In our implementation, we selected the candidate regulator with the highest BNS as a key regulator in each iteration. The number of key regulators was finalized based on the score distribution of BNS increase. Target genes were assigned to respective regulators under the constraint of a limited number of parents, which is denoted as d. For the first set of d regulators (parents for all target genes), all possible combinations of d regulators from the candidates were enumerated to select the combination that gave the highest BNS. Then, for a new regulator to be added, the parents of each target gene were combined to select the combinations of d regulators that gave the highest BNS. After summing all the BNSs, the candidate regulator that gave the highest score increase was added to the network. When selecting the (k + 1)th regulator, if the slope of the score increase was less than the threshold, the program stopped and selected the k regulators as the final parents of the target genes. The source codes and detailed instructions for searching the F. graminearum GRN can be accessed at: http://rio.ecs.umass.edu/Fusarium_GRN.
R GO score calculation
If regulators were SPs, functional enrichment using FunCat (Ruepp et al., 2004) was performed on target genes of each SP regulator to obtain significantly enriched terms with a P value threshold of 0.05. The significantly enriched terms were compared with functional terms of the SP regulator. The functional capture rate R GO was then calculated by dividing the number of shared functional terms by the total number of regulator functional terms.
Permutation test
A permutation test was performed to compare the R GO scores of our inferred regulatory network and the randomly generated regulator–target gene relationship. In each permutation, 120 regulators were randomly selected from candidate regulators. Each randomly selected regulator was mapped to the F. graminearum network with a set of randomly selected target genes of the same size. With the new random target genes, the R GO score for each random regulator was calculated following a FunCat analysis. After running permutation tests 100 times, the average distribution of the R GO score was compared with that of the inferred network.
Function prediction of TFs
Predicted TF regulators were evaluated using two different pipelines. First, the 75 F. graminearum TFs were searched for homologs in the S. cerevisiae genome (http://www.yeastgenome.org/) using Blastp (e < 1e‐10). For the identified F. graminearum TF homologs, FunCat analysis was performed for target genes and enriched functions were compared with S. cerevisiae TF homolog functions to identify consensus functions. Second, TF binding motif enrichment was performed for target genes of each F. graminearum TF homolog using degenerate TF binding motifs predicted previously (Kumar et al., 2010). The enriched TF binding motif was then compared with the binding motif in the S. cerevisiae TF homolog, if present in the YeTFaSCo database (http://yetfasco.ccbr.utoronto.ca), to identify similar binding motifs based on matching score computation.
TF binding site matching score
Generally, the TF binding motif is represented as a position weight matrix (PWM) or sequence logo. The motif matching score function was derived to measure the distance between the most enriched F. graminearum k‐mer motif and the known S. cerevisiae motif. The matching score for the most enriched motif m g and the published S. cerevisiae motif m y was calculated in Eqn 1:
| (Eqn 1) |
The PWM for S. cerevisiae motif m y is represented as a frequency matrix of M k,j, where k is one of possible nucleotides Σ = {A, C, G, T} and j is the position. The length of motif m y is denoted as m and the length of motif m g is denoted as n. The nucleotide at position i of m g is denoted as m g[i]. When m g is aligned with m y at position t of m y, the total matching score equals the summation of frequency M k,j at each aligned position. The denominator is the normalization factor to remove the influence of the maximum frequency. From all the possible aligned positions, the largest matching score is the m‐score.
Modularity analysis
Regulatory modules in the F. graminearum GRN were identified using the modularity function in Gephi (Bastian et al., 2009) employing randomization and a resolution score of 1.0. Genes of each regulatory module were exported and functional enrichment was conducted using FunCat. Gene expression levels of each module were extracted from master data tables using inhouse Python scripts, clustered and visualized using MEV (http://www.tm4.org/mev.html).
GRN–FPPI overlapping network analysis
To identify overlapping elements between the FPPI network and the GRN inferred in this study, genes and their interactions present in both networks were captured using inhouse Python scripts. FPPI network data were kindly provided by Professor Weihua Tang at the Institute of Plant Physiology and Ecology of the Chinese Academy of Sciences.
Statistical analysis
Statistical analyses, including Student's t‐test and Fisher's exact test, were conducted using the stats functions in the R programming language (http://www.r-project.org). The F. graminearum genome has been divided previously into core and species‐specific (FS) compartments containing c. 9700 and 3600 genes, respectively (Ma et al., 2010). For the top‐ranked 20 regulators for the two compartments, we performed Fisher's exact test using the fisher.test function in R. Our data provide strong support for the biased distribution of top regulators for both regions. Core regulators (19/20) are significantly enriched in the core genome compared with random chance (9700/13 300) (P = 0.02281), whereas FS regulators (1/20) are under‐represented in the core genome. Similarly, FS regulators (12/20) are significantly enriched in the FS genome compared with random chance (3600/13 300) (P = 0.002), whereas core regulators are significantly under‐represented in the FS genome.
Data access
Microarray data generated in this work using the Affymetrix Fungal Multigenome ExonChip (Guo et al., 2016) are available at the Filamentous Fungi Gene Expression Database (Zhang & Townsend, 2010) (Experiment ID: 241, 242, 246 and 247) (http://bioinfo.townsend.yale.edu/). The GRN inferred in this work, as well as related datasets, source codes and instructions for using the network, can be accessed at: http://rio.ecs.umass.edu/Fusarium_GRN.
Results
Global F. graminearum GRN inference
In this study, we inferred the F. graminearum GRN by adapting MinReg, a machine‐learning algorithm based on a BN model (Pe'er et al., 2002) which treats the expression level of each gene as a random variable and attempts to estimate the structural features of the dependences in their joint probability distribution from the data. To collect sufficient expression data for reliable network prediction, we generated 27 expression profiles based on nine experiments (see the Materials and Methods section) (Table S1) and acquired 166 transcriptomic datasets curated at the Plant Expression database (PLEXdb) (Dash et al., 2012). Expression data from both sources were combined and normalized, generating a global gene expression data matrix as input data (Table S2). A total of 968 candidate regulator genes, including 660 putative TFs and 308 SPs (Table S3), were identified based on current functional annotations and published studies (Cuomo et al., 2007; Wong et al., 2011). Overall, the global BNS increased when a new regulator was added (Fig. S2). At the beginning of the BNS distribution curve, there was a linear correlation between the global BNS value and the increase in regulators. The increase declined significantly after 120 regulators had been added. Theoretically, each candidate regulator can be fitted into the GRN; however, doing so is computationally costly and creates statistical noise which probably leads to spurious dependences with a relatively small sample size. Therefore, we terminated our search with a parsimonious set of major regulators and limited our search to a simple network structure in an effort to balance the high resolution of the learned networks with statistical robustness (see the Materials and Methods section). We defined these 120 regulators, which can be accessed at the website http://rio.ecs.umass.edu/Fusarium_GRN, as top regulators of the F. graminearum GRN (Fig. S3). The inferred top regulators included 75 TFs and 45 SPs. The number of target genes for each regulator ranged from 71 to 629, with an average of 329 (Fig. 1a). Because the expression data covered diverse biological states with an emphasis on pathogenesis, we aimed to deliver a reduced network structure that described the regulation of fundamental biological processes with an emphasis on the regulation of fungal pathogenesis. This framework can be further fitted with expression data under different perturbations to understand the regulation of specific biological functions.
Figure 1.
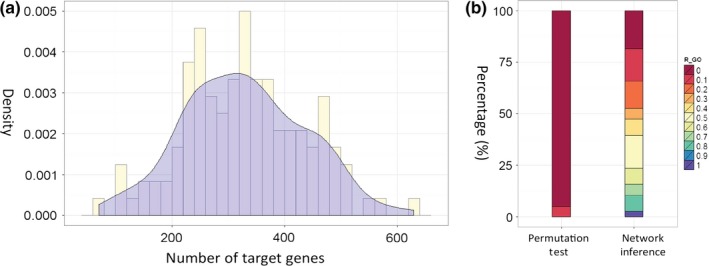
Summary of the Fusarium graminearum gene regulatory network (GRN). (a) The abundance of target genes for all regulators is summarized in a histogram and a density curve. x‐axis, number of target genes; y‐axis, density of regulators. (b) A stack bar plot comparing the R GO (function capture rate) distribution for signal protein (SP) regulators in the inferred GRN and the random network obtained by a permutation test (see the Materials and Methods section). R GO equals the ratio of functional terms shared by a regulator and its target genes (significantly enriched: P < 0.05) to the total functional terms of the regulator (see the Materials and Methods section). Color scale: R GO ranges from 0 to 1.
Validation of the predicted F. graminearum GRN
Top SP regulators share functions with their target genes
Regulators and their target genes are usually involved in the same biological processes (Pe'er et al., 2006). This is strongly supported by the top SP regulators identified in our study and the enriched gene ontology (GO) terms of their target genes. For example, 38 of the 45 inferred top SP regulators have been functionally annotated, which enables a comparison of the GO terms assigned to each regulator and its target genes using the ratio score function R GO (see the Materials and Methods section). The scores range from 0 to 1. A score of 1 reflects complete functional overlap, as all GO terms assigned to a regulator are enriched in the GO terms assigned to its target genes. By contrast, a score of 0 indicates a lack of functional overlap. Of the 38 annotated regulators, the average R GO score was 0.34, significantly higher than the randomized permutation test average R GO score of 0.0051 (Student's t‐test, P = 0.00205). Specifically, the R GO score for the regulator RAS2 (FGSG_10114) was 1, and six regulators had R GO scores of ≥ 0.7, and about half of the SP regulators (18) had R GO scores that were ≥ 0.4 (Fig. 1b; Table S4).
These top SP regulators, which act as controlling centers of the GRN and regulate many key processes essential for survival and proliferation, were found to be mostly involved in housekeeping cell functions, such as metabolism, cell cycle control, cellular transport, transcription, translation and cell growth (Table S4). For example, RAS2 (FGSG_10114) was predicted to regulate 557 target genes in the F. graminearum GRN. Empirically, the ras2 mutant showed multiple defects, including defects in vegetative growth, mating and virulence (Bluhm et al., 2007), even though the mutant was viable. Interestingly, these 557 target genes included 38 genes involved in the cell cycle, 41 in stress responses and 34 in cell polarity and ascospore development. Among the 45 SP regulators, at least three, that is, FGSG_00677 (subunit of casein kinase 2, CK2), FGSG_01137 (FgMPS1) and FGSG_09660 (FgPKC1), are essential (Fig. S4), and no viable knockout mutants could be experimentally obtained for these (Wang et al., 2011). Mutants of the other eight regulators showed phenotypic defects in one or more processes, including vegetative growth, sporulation, toxin production and virulence (Wang et al., 2011), supporting their involvement in essential cellular functions as top regulators (Fig. S4).
Most predicted top TF regulators are functionally conserved
Unlike SPs, most TFs in the F. graminearum genome have limited GO annotations associated with biological processes and lack functional annotations other than nucleic acid binding. Validation methods based on such incomplete information could amplify both false‐positive and false‐negative signals. Therefore, we took a homologous‐based approach, comparing the predicted top regulators with homologs in the best‐characterized model fungal genome, S. cerevisiae. Among the 75 predicted top TF regulators, 56 had putative S. cerevisiae homologs (e < 1e‐10). Transferring the functional annotation of yeast proteins to their F. graminearum homologs enabled us to perform an annotation‐based validation similar to that described for SPs. Interestingly, the functions of 34 of the 56 top TFs, as suggested by their yeast homologs, were confirmed by the enriched functional categories inferred by their target genes (see the Materials and Methods section) (Fig. 2a; Table 1). These 34 conserved regulators regulate a wide range of cellular processes, including the cell cycle, RNA processing during transcription and translation, mitochondrial functions, mating, stress responses and chromatin dynamics (Table 1).
Figure 2.
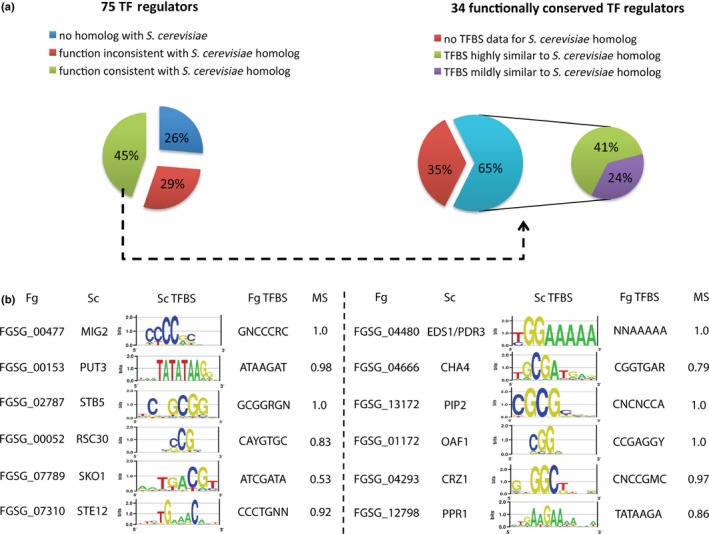
Transcription factor (TF) regulators inferred in the Fusarium graminearum gene regulatory network (GRN) have exceptional functional or TF binding site (TFBS) consistency with Saccharomyces cerevisiae homologs. (a) Pie charts summarizing the validation of 75 TF regulators inferred in the F. graminearum GRN compared with S. cerevisiae homologs in terms of function and TFBS. Dotted lines and arrows represent a further breakdown of functionally conserved TFs based on TFBS knowledge. (b) Summary of functionally conserved F. graminearum and S. cerevisiae TFs that share highly similar TFBSs, suggested by motif MSs (matching scores). MS computation details are provided in the Materials and Methods section.
Table 1.
Summary of conserved Fusarium graminearum (Fg) transcription factor (TF) regulators homologous to those in Saccharomyces cerevisiae (Sc)
| Fg TF regulators | Fg degenerate TFBS | Sc homologs | Sc TFBS evidence | Consensus function annotation | Motif matching score |
|---|---|---|---|---|---|
| FGSG_04666 | CGGTGAR | CHA4 | No | Cell cycle | –a |
| FGSG_07097 | CNCCAAN | YOX1 | Yes | Cell cycle control | 0.51 |
| FGSG_07924 | CGACNNC | ZNF1 | No | Cell cycle, cell component biosynthesis, cell differentiation, signal transduction | – |
| FGSG_07310 | CCCTGNN | STE12 | Yes | Cell cycle, cytokinesis, mating, cell type differentiation, stress response | 0.92 |
| FGSG_09807 | YGCGACN | CEF1 | No | Cell cycle, RNA processing, mRNA splicing | – |
| FGSG_08349 | NGTSACG | UME6 | Yes | Cell cycle, transcription, cell transport | 0.57 |
| FGSG_00385 | CCNCNTC | NHP6B | Yes | Chromatin remodeling, condensation | – |
| FGSG_08719 | NGCCNCA | MYO1 | No | Cytoplasmic and nuclear protein degradation, cell cycle, cytoskeleton structure | – |
| FGSG_07789 | ATCGATA | SKO1 | Yes | Disease, virulence, defense | 0.53 |
| FGSG_00477 | GNCCCRC | MIG2 | Yes | Fungal cell differentiation | 1 |
| FGSG_00318 | CCCCGSA | BAS1 | Yes | Metabolism, energy, transport | 0.71 |
| FGSG_00717 | AAAANTT | TIM10 | No | Mitochondria, mitochondria transport, heat shock stress response | – |
| FGSG_09892 | GATGNCN | MPE1 | No | mRNA synthesis, transcriptional control | – |
| FGSG_00153 | ATAAGAT | PUT3 | Yes | Nitrogen utilization | 0.98 |
| FGSG_04480 | NNAAAAA | EDS1 | Yes | Nonvesicular transport, stress response | 1 |
| FGSG_00800 | CCNCCNC | HAL9 | Yes | Osmotic and salt stress response, stress response | 1 |
| FGSG_13172 | CNCNCCA | PIP2 | Yes | Oxidation of fatty acids, cell cycle and DNA processing | 1 |
| FGSG_06516 | GCKGACT | STB5 | Yes | Oxygen and radical detoxification | 1 |
| FGSG_01172 | CCGAGGY | OAF1 | Yes | Peroxisome, fatty acid, mitochondria | 1 |
| FGSG_01411 | NNCGCGT | PFA4 | No | Protein modification, degradation, cell cycle regulation | – |
| FGSG_06220 | GNGGGGY | SUI2 | No | Protein synthesis, translation, translation initiation | – |
| FGSG_05012 | AGCTNCN | ARC1 | No | Protein synthesis, tRNA aminoacylation, tRNA binding, stress response | – |
| FGSG_00052 | CAYGTGC | RSC30 | Yes | Regulation of ribosomal protein genes | 0.83 |
| FGSG_04554 | YNATTGG | DPS1 | No | RNA modification, rRNA processing | – |
| FGSG_06168 | RAAAAAN | RSF2 | Yes | rRNA processing, protein synthesis, transcription | 0.84 |
| FGSG_11996 | ACGTMAT | ECM22 | Yes | Sterol metabolism | 0.62 |
| FGSG_00713 | NCTCCCN | STB5 | Yes | Stress response | 0.61 |
| FGSG_12798 | TATAAGA | PPR1 | Yes | Stress response | 0.86 |
| FGSG_02696 | NACGTCA | PPR1 | Yes | Stress response | 0.84 |
| FGSG_02787 | GCGGRGN | STB5 | Yes | Stress response | 1 |
| FGSG_04293 | CNCCGMC | CRZ1 | Yes | Stress response, calcium binding | 0.97 |
| FGSG_01638 | GCGNCAN | HAP1 | Yes | Transcription regulation | 0.64 |
| FGSG_00324 | CNCCCNC | SNT1 | No | Transcription, chromatin regulation, meiosis | – |
| FGSG_05498 | GANGCGN | BUR6 | No | Transcription, chromosomal cycle | – |
TFBS, transcription factor binding site.
Score not calculated.
A comprehensive mutagenesis study generated and characterized all 660 TFs in the F. graminearum genome, showing that 170 TF deletion mutants have at least one phenotypic defect (Son et al., 2011), including 13 of the 75 predicted top TF regulators (Fig. S4). Interestingly, 10 of these 13 TFs were found to be conserved in S. cerevisiae. For example, FGSG_07310 was found to be the homolog of S. cerevisiae STE12, which is activated by the MAPK cascade in response to pheromones and controls mating (Elion et al., 1993). The FGSG_07310 deletion mutant is unable to reproduce sexually, confirming the essential role of FGSG_07310 in mating (Son et al., 2011). Deletion mutants of four of these TFs, FGSG_00324, FGSG_00385, FGSG_00477 and FGSG_08719 (Fig. S4), had multiple defects (Son et al., 2011), consistent with the fundamental biological processes they share with their S. cerevisiae homologs, such as transcription, and chromatin and cytoskeleton structure (Table 1).
TFs control the transcription of their target genes by binding to their respective regulatory sequences, called TFBSs, at the promoter regions of the target genes. It has been reported repeatedly that the sequence of a TFBS can be preserved in a conserved TF over evolutionary distance, for instance from unicellular yeasts to filamentous fungi (Kumar et al., 2010; Wittkopp & Kalay, 2012). Based on the set of computationally predicted TF cis elements in the F. graminearum genome (Kumar et al., 2010), we identified the most enriched TF binding sites (Student's t‐test) among target genes for each inferred top TF regulator (Table S5). On average, about one‐quarter of the predicted target genes of each TF shared an enriched binding site. We further developed a score function based on PWM to measure the sequence similarity between the enriched F. graminearum binding motifs and the corresponding ones predicted in the Yeast Transcription Factor Specificity Compendium (see the Materials and Methods section). The score ranges from 0 to 1, representing the least to the most matches. Among the 34 functionally conserved F. graminearum–S. cerevisiae TFs, 22 (65%) shared TF binding sequences with S. cerevisiae homologs (Fig. 2a; Table 1). The conserved binding motifs included the TFBS for MIG2, PUT3, EDS1, STE12 and STB5, with matching scores of higher than 0.8 (Fig. 2b).
In summary, validation of the reduced F. graminearum GRN confirms its reliability in depicting the relationship between top regulators, both SPs and TFs, and their target genes. The conservation between F. graminearum and S. cerevisiae for both the predicted top TFs and their TFBSs suggests that most predicted top regulators in this reduced network structure perform conserved housekeeping functions.
Modularity is a key feature of the F. graminearum GRN
The GRN usually contains interconnected functional modules that have specific functions in biological processes (Segal et al., 2003). Multiple software packages, such as Gephi (Bastian et al., 2009), were designed to extract tightly connected subnetworks from larger networks (Blondel et al., 2008), including social networks such as Facebook (Akhtar et al., 2013), as well as transcription regulatory networks (Sanz et al., 2011). Using Gephi, the F. graminearum GRN can be reproducibly divided into eight regulatory modules (Fig. 3). GO term enrichment tests (Table S6) confirmed that each module was functionally independent and was important for certain biological functions (Fig. 4). However, some functions were controlled by multiple modules (Tables 2, S6). For example, module A was found to function in the cell cycle, remodeling of chromatin structure and cytoskeletal organization, and cell polarity. Modules D, E and F were found to regulate translation, transcription and DNA replication, respectively. Module G was found to control cell differentiation and stress responses. Although modules B, C and H were all found to be involved in the regulation of cell detoxification processes, they were enriched for different cellular processes. For instance, module B was involved in secondary metabolism, including lipid, fatty acid and isoprenoid metabolism; module C was highly enriched for cytochrome P450‐related detoxification processes; and module H primarily regulated carbohydrate metabolism and detoxification.
Figure 3.
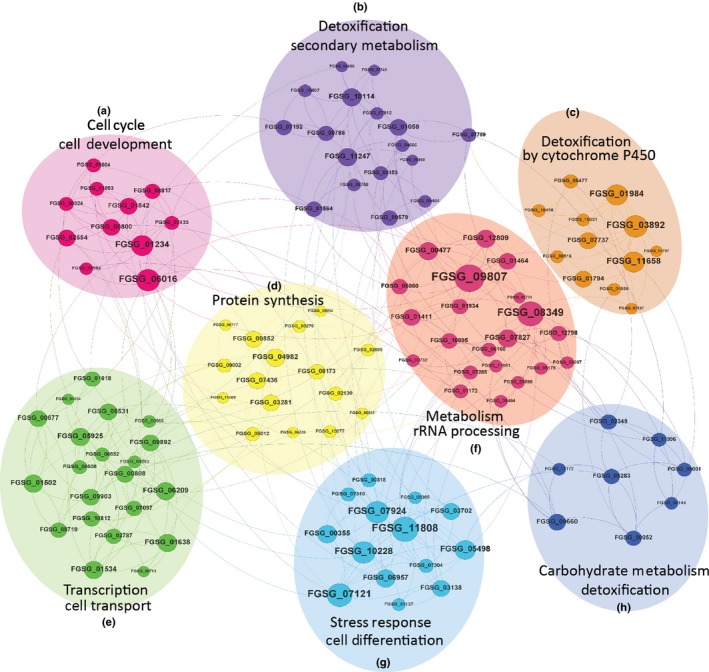
The Fusarium graminearum gene regulatory network (GRN) consists of eight regulatory modules that are regulated under different biological states. Visualization of the F. graminearum gene regulatory network, which is divided into eight modules (a–h). Only the regulator nodes are shown. Modules (shaded circles) and their enclosed regulator nodes are colored. The size and label of the nodes are proportional to the number of target genes for each regulator. Edges are nonweighted, simply showing inferred regulatory relationships. The functional annotation of each module represents the most significantly enriched gene ontology terms among target genes within the module.
Figure 4.
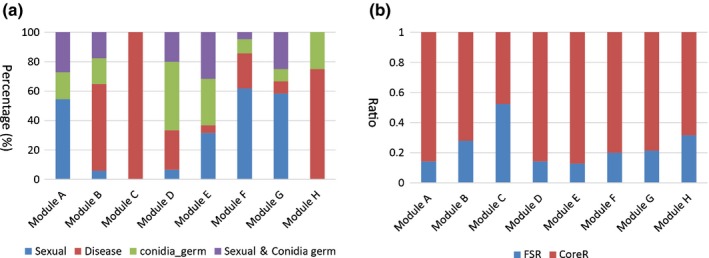
Dissection of functional association in regulatory modules of Fusarium graminearum. (a) Bar graphs summarizing the number of regulators belonging to the four major regulator clusters: Sexual, Disease, conidia_germ (conidia germination) and Sexual&Conidia germ, equivalent to the clusters shown in Fig. 3. The regulators were counted for each module and plotted. Bar colors represent the four clusters. (b) Core genome and F. graminearum‐specific distribution of regulators for each regulatory module. FSR, F. graminearum‐specific genome regulators; CoreR, core genome regulators.
Table 2.
Summary of module statistics: the number of target genes and regulators in each module and the most significantly enriched functional terms in target genes ranked by P values (low to high)
| Module | No. target genes | No. FS target genes (percentage)a | FS enrichment (P value)b | No. regulators | Significantly enriched biological processesc |
|---|---|---|---|---|---|
| A | 2743 | 367 (13.4) | Under (5.4e‐38) | 11 | Cell cycle; cytoskeleton; budding; cell polarity; chromosomal structure; G‐protein‐mediated cell signaling |
| B | 5066 | 1539 (30.4) | Over (1.1e‐3) | 17 | Detoxification; degradation of exogenous compounds; nonvesicular cellular import; secondary metabolism |
| C | 3195 | 1658 (52) | Over (1.3e‐71) | 12 | Degradation of exogenous compounds; detoxification involving cytochrome P450; polysaccharide metabolism |
| D | 3295 | 594 (18) | Under (3.4e‐18) | 16 | Protein synthesis; unfolded protein response; mitochondrion; stress response |
| E | 4440 | 561 (12.6) | Under (3.8e‐63) | 21 | Transcription; cellular transport; cytoskeleton; cell cycle; mitochondrion; stress response |
| F | 6807 | 1693 (24.8) | Under (8.4e‐3) | 21 | Metabolism; rRNA processing; Ori recognition and priming complex formation; vitamin/cofactor transport |
| G | 4212 | 861 (20.4) | Under (6.5e‐12) | 14 | Stress response; fungal cell type differentiation; ascospore development; cell wall; G‐protein‐mediated signal transduction |
| H | 3012 | 1151 (38.2) | Over (1.2e‐17) | 8 | Carbohydrate metabolism; detoxification by degradation; aminosaccharide catabolism |
FS, Fusarium graminearum specific. Percentage, FS target gene ratio per module.
FS enrichment: enrichment test (two‐sided Fisher's exact test) for FS target genes over total target genes per module, compared with total FS genes (3600) against whole genome (13 300). Over, over‐representation or enrichment (P < 0.05). Under, under‐representation or lack of enrichment (P < 0.05).
Significantly enriched biological processes using FunCat database (ranked by FunCat P‐value in ascending order). Complete FunCat results for all modules are available in Supporting Information Table S6.
Such functional affiliation was also supported by the findings that most regulators and target genes in modules B, C and H were upregulated during plant infection (Fig. S5) and that the gene expression profiles of modules B, C and H were distinct from those of the other modules (Fig. S5; Table 2). For instance, module A (cell cycle and cell development module) was preferentially expressed during sexual reproduction stages, but module D (translation module) was induced during conidial germination processes (Fig. S5). To quantify the functional affiliation of each module, we co‐clustered the expressed profiles of the 120 top regulators and the expression profiles used for the F. graminearum GRN inference employing a hierarchical clustering (HCL) algorithm (Fig. S6), and observed distinct clustering patterns for the regulatory modules (Fig. 4a), in agreement with their functional annotations. The most significant affiliation was in module C, where all regulators were found to be related to the disease phenotype (Fig. 4a) and were strongly induced during plant infection (Fig. S5). In addition to module C, modules B (57% of regulators) and H (70%) were also substantially correlated with disease phenotypes. Of 22 candidate FS effectors reported previously (Brown et al., 2012), 12, 12 and eight were found in modules B, C and H, respectively. Although all modules contain effector genes, B and C have the highest ratio of effector genes per module (Table S7). By contrast, modules A (80%), F (68%) and G (85%) were major contributors to sexual development. Modules D (70%) and E (68%) were primary contributors to the conidial germination process (Fig. 4a). This distinctiveness of expression profiles and functional affiliations among different modules indicates that the F. graminearum GRN has clear modularity and that each module is differentially expressed under certain biological conditions.
The F. graminearum GRN reveals conserved vs species‐specific components
Novel traits, such as overcoming host resistance to establish infection, or utilizing different nutrient sources to support growth in a changing environment, are important for the adaptation of an organism and, in many cases, are gained through the acquisition of new genes. The integration of these new genes into the regulatory network is crucial for their functionality. Based on a knowledge of the structural and functional divergence of the F. graminearum genome (Cuomo et al., 2007), we identified 9700 orthologous genes that were conserved among three Fusarium sister species, F. graminearum, F. verticillioides and F. oxysporum (Ma et al., 2010), as core genes. About 3600 genes that lack orthologous sequences in the sister species were loosely defined as F. graminearum specific (FS hereafter). These FS genes were primarily localized to genomic regions that contained a high density of single nucleotide polymorphisms (SNPs), and some of these genes were found to encode pathogenicity‐related effector proteins (Kistler et al., 2013) and secondary metabolite gene clusters (Zhao et al., 2014).
Based on this genome partition, we examined whether the regulatory modules identified in the F. graminearum GRN had an enrichment of genes located on either genomic region. Interestingly, FS genes are significantly enriched in the three pathogenesis‐related modules, B (30%, P = 1.1e‐3), C (52%, P = 1.3e‐71) and H (38.2%, P = 1.2e‐17), but under‐represented in all other modules (Table 2). Such a biased distribution is reflected in both regulators (Fig. 4b) and their target genes, and the most significant enrichments are observed among FS target genes that are regulated by FS regulators. For instance, 55% of FS target genes in module B, 84% of FS target genes in module C and 73% of FS target genes in module H are regulated by FS regulators.
To further understand the compartmentalization of regulation, we ranked the top regulators based on their number of target genes, and searched for the top 20 regulators for the conserved vs species‐specific genes, respectively. For the 9700 core genes, 19 of the 20 top regulators were also encoded in the core regions, indicating significant enrichment if assuming a random distribution (P = 0.02281). By contrast, only eight of the highest ranked 20 regulators for the 3600 FS genes were found to be core regulators, and 12 were FS regulators (Fig. 5a), which were significantly enriched (P = 0.002) (see the Materials and Methods section). This over‐representation of core and FS regulators for the core and FS genomes, respectively, suggests that each genomic region has a regulatory network that primarily employs regulators from the same region. In addition, gene expression patterns for the top 20 regulators of the two genomic regions were also distinct. In contrast with the top regulators in the core genome, those in the FS genome were highly upregulated during plant infection stages, further suggesting that the FS genome and its top regulators play vital roles in fungal pathogenesis (Fig. 5a).
Figure 5.
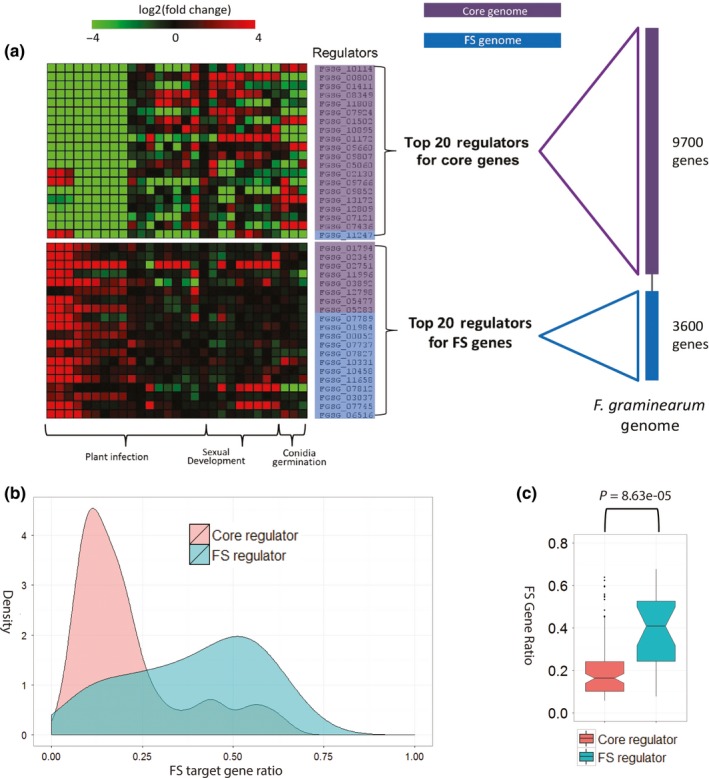
The Fusarium graminearum gene regulatory network (GRN) has core genome‐ and F. graminearum‐specific (FS) components. (a) Top 20 regulators (in terms of having the most target genes) of target genes belonging to the core genome and FS genome. Purple and blue rectangles denote the core genome and FS genome, respectively. The same colors were also used to shade the top 20 regulators, based on their location in genome compartments. Heatmaps show the gene expression changes of the 40 regulators across three categories of biological conditions. Color scale represents log2‐transformed fold change from low (−4) to high (4). Green, downregulated; red, upregulated; black, no change. The core and FS genomes were defined based on a comparative genome analysis between F. graminearum and three additional Fusarium species: F. oxysporum f. sp. lycopercisi, F. verticillioides and F. solani. Core genes are shared by the four species. FS genes are unique to F. graminearum. (b) Kernel density curve of FS target gene ratio distributions for 96 core and 24 FS regulators in the F. graminearum GRN. FS target gene ratios measure the proportion of FS genes amongst all target genes per regulator. (c) Notch boxplots summarizing FS target gene ratio distributions for 96 core and 24 FS regulators, which are significantly different, with P < 0.05 (Student's t‐test). A horizontal black line across each box denotes the mean gene ratio.
Furthermore, we found that if a regulator was F. graminearum specific, its target genes were mostly F. graminearum specific, and vice versa. We calculated the core and FS target gene ratios, which represent the frequency of target genes from the core or FS genome, respectively, in the total number of target genes per regulator. Density curves suggested that 96 core regulators (regulators from the core genome) had low FS target gene ratios, compared with 24 FS regulators (Fig. 5b). The FS gene ratio distributions for 24 FS regulators (mean, 0.41) and 96 core regulators (mean, 0.18) were significantly different (Student's t‐test: P = 8.63e‐05) (Fig. 5c). By contrast, target genes inferred for core regulators were mostly situated in the core genome, and the FS regulators had low core target gene ratios (Fig. S7). These findings indicate that a compartmentalization of regulatory networks exists between the core and FS genomic regions.
A comparison of the F. graminearum GRN and FPPI reveals subnetworks
Many regulatory interactions are effectively carried out by protein complexes, which are essential molecular machineries for a vast array of biological functions. FPPI, which interconnects 3750 proteins, was established previously (Zhao et al., 2009). By comparing the F. graminearum GRN with the reported FPPI, we identified shared components that capture a total of 311 interactions between 337 proteins. Although this shared network contained only 337 proteins, 78 were predicted top regulators, including 34 SP regulators and 44 TF regulators identified in this study (Fig. S8; Table S8).
These shared interactions formed several major subnetworks, and the top two were a ribosomal protein complex and a complex that involved a subunit of CK2 (FGSG_00677). The ribosomal protein complex contained 30 proteins that were all annotated as ribosomal protein subunits and predicted to be part of module D, which was found to be involved in translation. The subnetwork centering on the regulator FGSG_00677 included a total of 20 proteins and was part of module E, which was found to control transcription and cell transport. FGSG_00677, an essential F. graminearum kinase (Wang et al., 2011), was found to be involved in cell cycle control, rRNA synthesis, and cell growth and polarity (Table S4). Overall, we found that proteins involved in the cell cycle and transcription were enriched in this subnetwork. Consistent with the active involvement of protein synthesis, DNA synthesis and transcription during fungal spore germination (Osherov & May, 2001; Seong et al., 2008), both modules D and E were strongly transcriptionally associated with conidial germination (Fig. 4a).
The other shared component had a key regulator, adenylate cyclase FAC1 (FGSG_01234), which controls the central signaling cascade cyclic adenosine monophosphate–protein kinase A (cAMP–PKA) pathway by producing the second messenger cAMP (D'Souza & Heitman, 2001) to transmit extracellular stimuli and govern cell responses (Hu et al., 2014; Guo et al., 2016). This component was part of module A in the F. graminearum GRN and was predicted to regulate the cell cycle and cell development.
Previously, our comparative study of the cAMP–PKA pathway in F. graminearum and in one of its sister species, F. verticillioides (Guo et al., 2016), has revealed that this pathway has conserved housekeeping functions, such as cell cycle regulation, primary metabolism and stress response. In the F. graminearum GRN, FAC1 was predicted to be one of the top regulators, and was found to regulate 356 target genes that were enriched for functions in small GTPase‐mediated signaling, cell cycle regulation and ascospore development (Table S4). Combining FPPI, our GRN inference and the 1238 differentially expressed genes (DEGs) identified in the Δfac1 mutant (Guo et al., 2016), we identified a single edge that was supported by all three lines of evidence (DEG–GRN–FPPI) (Fig. 6a,b). This single node (FGSG_09908) encoded the regulatory subunit of PK (PKR), and binding of cAMP to this subunit led to activation of CPK1. Most edges were supported by both GRN and DEG (26 edges), including FGSG_00800 (HAL9) and FGSG_08763 (SWI6). Seven edges were supported by GRN and FPPI, including a RAS GTPase (FGSG_05501) and protein phosphatases PP2, PP2c and CDC25. Interestingly, 16 edges were supported by DEG and FPPI, but not by GRN prediction, including a FAC1‐associated protein (FGSG_01923), the cAMP scavenger enzyme PDE1 (FGSG_06633), G‐protein β subunit FGB1 (FGSG_09870) and a MAPK (FGSG_09612). This suggests that these proteins may be part of a large protein complex involved in the cAMP–PKA signaling pathway, but may not be directly regulated by FAC1 (Fig. 6b).
Figure 6.
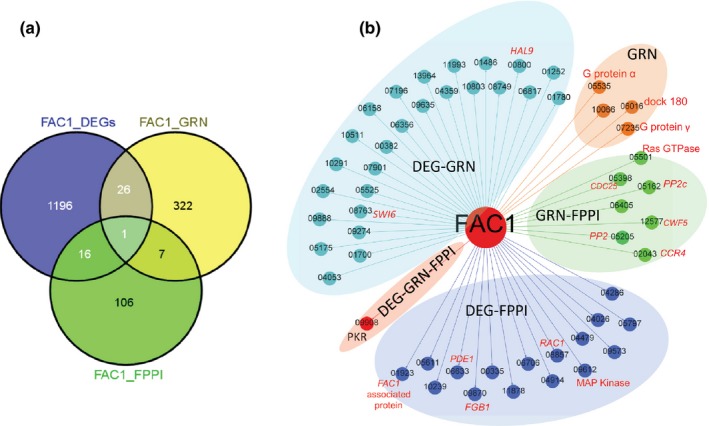
Fusarium graminearum FAC1 regulatory subnetworks. (a) Venn diagram of network edges captured by the gene regulatory network (GRN), F. graminearum protein–protein interaction network (FPPI) and differentially expressed genes (DEG) for the ∆fac1 mutant. (b) Fusarium graminearum FAC1 (FGSG_01234, adenylate cyclase) regulatory subnetworks. Edges are nonweighted and depict regulatory relationships. For simplicity, prefixes (FGSG_) of genes are omitted. Five clusters within the network are: GRN shared with DEG (DEG‐GRN), GRN shared with FPPI (GRN‐FPPI), FPPI shared with DEG (DEG‐FPPI), shared by all three (DEG‐GRN‐FPPI) and only in GRN (all are not shown). Colors of nodes (except FAC1, the regulator) are the same as the clusters. Red labels beside the nodes are either gene names or functional annotations of genes.
Because this current model links all target genes with a subset of regulators, we anticipate a relatively high false‐positive rate in target prediction. Therefore, it is not surprising that we identified over 300 targets that only existed in GRN and lacked support from either DEG or FPPI. These target genes had multiple regulators in GRN; therefore, it is likely that their expression levels may not be changed by blocking a single regulator. However, the GRN‐unique edges did capture cAMP signaling‐related genes, such as the G‐protein α subunit (FGSG_05535) and γ subunit (FGSG_07235), consistent with the fact that G‐protein signaling regulates cAMP signaling. FAC1 is likely to be co‐regulated with these target genes.
Overall, the network analysis reported here captures many edges that can be supported by both protein–protein interaction and gene expression profiles. Combining these multiple layers of evidence helps us to understand the hierarchical structure of the FAC1 regulatory network in depth. This analysis provides a template for functional validation of GRN, which could be achieved using genetic and physical interaction data obtained in future biological experiments.
Discussion
The reconstruction of a GRN at the genomic level remains a daunting task in any organism, particularly in higher eukaryotes. In this study, we selected a filamentous fungus, F. graminearum, for the network inference, because of its agricultural importance, the wealth of available transcriptomic data and the existence of a well‐annotated genome. Furthermore, F. graminearum is one of the most extensively researched plant‐pathogenic fungi (Dean et al., 2012). This first reconstructed GRN of a filamentous ascomycete identified 120 top regulators that regulate different sets of target genes, and provided a regulatory framework that links every gene in the genome. Phenotypic association analysis showed that some of the identified top regulators are critical for fungal asexual and sexual growth, toxin production and virulence. Interestingly, although the SP regulators investigated to date seem to be crucial or indispensable, most TF regulator mutants have similar phenotypes to the wild‐type, with a few exceptions. This may be attributed to different modes of action for these two different types of regulator. SPs, such as kinases, typically act upstream of TFs and are therefore higher on the hierarchy of regulators, behaving as molecular switches. However, TFs often require the cooperation of multiple transcriptional cofactors, as this perhaps allows cells to cope with the loss of certain individual TFs. Regardless, the importance of kinases vs TFs is not always intuitive and, in many cases, requires further investigation.
One significant discovery of this relatively preliminary network inference is the modularity of the network. Any biological function is accomplished by hundreds of proteins through independent, but interconnected, subnetworks. Even though complete separation may be impossible, certain subnetworks are more interconnected than others. Our modularity analysis reproducibly identified eight regulatory modules. Although this distinction between modules is clear, it is not uncommon to find genes associated with more than one module, reflecting the complexity of regulatory networks. The study of these modules will provide valuable insight into the gene regulatory circuits that control key fungal biological processes, and guide the functional characterization of these regulators and target genes through experimental approaches.
The other intriguing discovery of this study is the compartmentalization of the regulatory network. Previous studies have divided the F. graminearum genome into conserved vs highly variable genomic regions. Genes encoded within these regions are enriched for species‐specific genes and many of these encode potential virulence factors that are induced during plant infection (Cuomo et al., 2007; Zhao et al., 2014). In agreement with the genomic division, these two regions are controlled by compartmentalized regulatory circuits in the predicted F. graminearum GRN. However, there are strong connections between these two regions and they clearly act as one unit. How F. graminearum acquired these novel regulatory circuits during evolutionary processes, how they were integrated with core circuits to form a cohesive functional unit and, finally, how the two parts communicate to regulate diverse fungal biological processes remain to be determined.
Our network inference also provides insight into the TF binding sites for the predicted TF regulators using a purely computational approach. Based on previously published results of an in silico cis‐element prediction for F. graminearum (Kumar et al., 2010) and on homology with S. cerevisiae, we found strong conservation signals for 12 TFs (of 75 predicted top TF regulators) and their corresponding TFBSs (matching score > 0.8) between F. graminearum and S. cerevisiae, indicating functional conservation over > 400 million yr of evolutionary time. As expected, we observed sequence divergence for the binding sites, even when the TFs were conserved, as TF binding site turnover is commonly observed during fungal species evolution, even between closely related species (Gasch et al., 2004).
Overall, this network prediction illustrates the power of computational biology and demonstrates the integration of social network and life sciences. However, this current static network model has its limitations. First, it does not consider the dynamic nature of the network and lacks the fine resolution needed to identify regulatory relationships under specific conditions. For example, two well‐characterized TFs, Tri6 and Tri10, which control the production of the mycotoxin trichothecene, are absent in the top regulator list. This is probably a result of their specialized, but nonessential, regulation of a set of genes that control secondary metabolism. Indeed, when we extended our search, both Tri6 and Tri10 were identified among the top 500 regulators. Second, the current algorithm is based on the consistent correlation in expression levels of a regulator and target genes without any consideration of post‐transcriptional and post‐translational regulation events, which may have created noise in the prediction. Third, certain genes that share similar patterns of expression without actual regulatory relationships may have been assigned regulatory relationships as false positives. As expression‐based modeling identifies regulators that are active (up‐ or downregulated) under most, if not all conditions, candidate regulators lacking significant expression changes are unlikely to be identified. Despite these limitations, we are confident of the overall accuracy of this network model, which has already revealed exciting and biologically meaningful insights into the gene regulation of this filamentous fungus. By increasing the number of datasets obtained under more diverse conditions, the network structure and topology can be further refined with improved resolution. The identification of the components and dynamics of the cell regulatory network and the pinpointing of specific regulators that govern fungal pathogenesis will offer potential novel strategies to control FHB, one of the most devastating diseases of wheat.
Author contributions
The project was designed by L. Guo, G.Z., L. Gao and L‐J.M. The manuscript was written by L. Guo, G.Z., L. Gao and L‐J.M. The experimental design was conducted by L. Guo, G.Z., H.C.K., J‐R.X. and L‐J.M. Chip design was conducted by H.C.K., J‐R.X. and L‐J.M.
Supporting information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Scatterplot matrix of three biological replicates and associated Pearson correlation coefficients (r) for each condition/biological state transcriptomically profiled in this study using the Affymetrix Fungal Multigenome ExonChip.
Fig. S2 Bayesian network score distribution for the top 300 regulators in the Fusarium graminearum gene regulatory network (GRN).
Fig. S3 Visualization of the Fusarium graminearum gene regulatory network (GRN) featuring the top 120 regulators, including transcription factors (TFs) and signal proteins (SPs), represented by red and green nodes, respectively.
Fig. S4 Gene–phenotype networks depicting the association of known phenotypes (red nodes) with inferred signal protein (SP) (blue nodes) and transcription factor (TF) (green nodes) regulators.
Fig. S5 Regulatory modules are differentially regulated in response to various biological conditions.
Fig. S6 Heatmap of hierarchical co‐clustering of biological conditions (rows) and 120 regulators (columns).
Fig. S7 Biased regulatory networks for core regulators of Fusarium graminearum.
Fig. S8 Shared networks by the Fusarium graminearum gene regulatory network (GRN) and protein–protein interaction network (FPPI).
Table S1 Summary of conditions on which the transcriptomic data are based for Fusarium graminearum gene regulatory network inference in this study
Table S4 Summary of signal proteins identified as inferred regulators in this network
Table S2 Input gene expression data matrix
Table S5 List of 75 inferred transcription factor regulators and the most enriched binding sites for target genes
Table S8 Shared networks by the Fusarium graminearum gene regulatory network (GRN) and protein–protein interaction network (FPPI)
Table S3 Spreadsheets containing lists of candidate regulators, inferred 120 top regulators, the full gene regulatory network (GRN), core genome GRN and Fusarium graminearum‐specific GRN
Table S6 Gene lists of eight regulatory modules (A–H) and complete FunCat analysis results
Table S7 Gene regulatory networks for candidate effector genes
Acknowledgements
This project was supported by the National Research Initiative Competitive Grants Program grant nos. 2008‐35604‐18800 and MASR‐2009‐04374 from the USDA National Institute of Food and Agriculture. L. Guo and L‐J.M. were also supported by a seed grant from Massachusetts Green High Performance Computing Center and the National Research Initiative Hatch Grants Program grant no. MAS00441. The authors would like to extend their thanks to Steve Klosterman, Lynda Ciuffitti, Ralph Dean and Andrew Breakspear for providing DNA and RNA samples; to Supriya Gupta and Andrew Crenshaw at the Broad Institute Expression Platform for conducting hybridization of both DNA and RNA samples to the Affymetrix arrays and for processing the microarray results; and to Gene Tanimoto and Stan Trask at Affymetrix for their assistance with the design of the fungal pathogen microarrays and for optimizing the hybridization conditions.
References
- Akhtar N, Javed H, Sengar G. 2013. Analysis of Facebook social network. IEEE (Institute of Electrical and Electronics Engineers) International Conference on Computational Intelligence and Computer Networks (CICN), 27–29 September 2013, Mathura, India. Institute of Electrical and Electronics Engineers, 451–454.
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O et al 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M 2009. Gephi: an open source software for exploring and manipulating networks In: Adar E, Hurst M, Finin T, Glance NS, Nicolov N, Tseng BL, eds. The international conference on weblogs and social media. Palo Alto, CA, USA: The AAAI Press, 361–362. [Google Scholar]
- Blondel VD, Guillaume J‐L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. Journal of Statistical Mechanics 10: 10008. [Google Scholar]
- Bluhm BH, Zhao X, Flaherty JE, Xu JR, Dunkle LD. 2007. RAS2 regulates growth and pathogenesis in Fusarium graminearum . Molecular Plant–Microbe Interactions 20: 627–636. [DOI] [PubMed] [Google Scholar]
- Boddu J, Cho S, Kruger WM, Muehlbauer GJ. 2006. Transcriptome analysis of the barley–Fusarium graminearum interaction. Molecular Plant–Microbe Interactions 19: 407–417. [DOI] [PubMed] [Google Scholar]
- Brown NA, Antoniw J, Hammond‐Kosack KE. 2012. The predicted secretome of the plant pathogenic fungus Fusarium graminearum: a refined comparative analysis. PLoS ONE 7: e33731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuomo CA, Guldener U, Xu JR, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma LJ, Baker SE, Rep M et al 2007. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science 317: 1400–1402. [DOI] [PubMed] [Google Scholar]
- Darabos C, Di Cunto F, Tomassini M, Moore JH, Provero P, Giacobini M. 2011. Additive functions in boolean models of gene regulatory network modules. PLoS ONE 6: e25110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Van Hemert J, Hong L, Wise RP, Dickerson JA. 2012. PLEXdb: gene expression resources for plants and plant pathogens. Nucleic Acids Research 40: D1194–D1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R, Van Kan JA, Pretorius ZA, Hammond‐Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J et al 2012. The Top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology 13: 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza CA, Heitman J. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiology Reviews 25: 349–364. [DOI] [PubMed] [Google Scholar]
- Elion EA, Satterberg B, Kranz JE. 1993. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Molecular Biology of the Cell 4: 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe'er D. 2000. Using Bayesian networks to analyze expression data. Journal of Computational Biology 7: 601–620. [DOI] [PubMed] [Google Scholar]
- Gardiner DM, Kazan K, Manners JM. 2009. Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Molecular Plant–Microbe Interactions 22: 1588–1600. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Moses AM, Chiang DY, Fraser HB, Berardini M, Eisen MB. 2004. Conservation and evolution of Cis‐regulatory systems in ascomycete fungi. PLoS Biology 2: e398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass L, Kauffman SA. 1973. The logical analysis of continuous, non‐linear biochemical control networks. Journal of Theoretical Biology 39: 103–129. [DOI] [PubMed] [Google Scholar]
- Goswami RS, Kistler HC. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Molecular Plant Pathology 5: 515–525. [DOI] [PubMed] [Google Scholar]
- Guelzim N, Bottani S, Bourgine P, Kepes F. 2002. Topological and causal structure of the yeast transcriptional regulatory network. Nature Genetics 31: 60–63. [DOI] [PubMed] [Google Scholar]
- Guldener U, Mannhaupt G, Munsterkotter M, Haase D, Oesterheld M, Stumpflen V, Mewes HW, Adam G. 2006a. FGDB: a comprehensive fungal genome resource on the plant pathogen Fusarium graminearum . Nucleic Acids Research 34: D456–D458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldener U, Seong KY, Boddu J, Cho S, Trail F, Xu JR, Adam G, Mewes HW, Muehlbauer GJ, Kistler HC. 2006b. Development of a Fusarium graminearum Affymetrix GeneChip for profiling fungal gene expression in vitro and in planta . Fungal Genetics and Biology 43: 316–325. [DOI] [PubMed] [Google Scholar]
- Guo L, Breakspear A, Zhao G, Gao L, Kistler HC, Xu J‐R, Ma L‐J. 2016. Conservation and divergence of the cyclic adenosine monophosphate–protein kinase A (cAMP–PKA) pathway in two plant‐pathogenic fungi: Fusarium graminearum and F. verticillioides . Molecular Plant Pathology 17: 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Ma L‐J. 2014. Fusarium graminearum genomics and beyond In: Dean RA, Lichens‐Park A, Kole C, eds. Genomics of plant‐associated fungi: monocot pathogens. Berlin/Heidelberg, Germany: Springer, 103–122. [Google Scholar]
- Hallen HE, Huebner M, Shiu SH, Guldener U, Trail F. 2007. Gene expression shifts during perithecium development in Gibberella zeae (anamorph Fusarium graminearum), with particular emphasis on ion transport proteins. Fungal Genetics and Biology 44: 1146–1156. [DOI] [PubMed] [Google Scholar]
- Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genetics 5: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Zhou X, Gu X, Cao S, Wang C, Xu J‐R. 2014. The cAMP‐PKA pathway regulates growth, sexual and asexual differentiation, and pathogenesis in Fusarium graminearum . Molecular Plant–Microbe Interactions 27: 557–566. [DOI] [PubMed] [Google Scholar]
- Hu Z, Killion PJ, Iyer VR. 2007. Genetic reconstruction of a functional transcriptional regulatory network. Nature Genetics 39: 683–687. [DOI] [PubMed] [Google Scholar]
- Jenczmionka NJ, Schafer W. 2005. The Gpmk1 MAP kinase of Fusarium graminearum regulates the induction of specific secreted enzymes. Current Genetics 47: 29–36. [DOI] [PubMed] [Google Scholar]
- Jonkers W, Dong Y, Broz K, Kistler HC. 2012. The Wor1‐like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum . PLoS Pathogens 8: e1002724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman SA. 1969. Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of Theoretical Biology 22: 437–467. [DOI] [PubMed] [Google Scholar]
- Kim H, Hu W, Kluger Y. 2006. Unraveling condition specific gene transcriptional regulatory networks in Saccharomyces cerevisiae . BMC Bioinformatics 7: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HD, Shay T, O'Shea EK, Regev A. 2009. Transcriptional regulatory circuits: predicting numbers from alphabets. Science 325: 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Waddell PJ. 2000. Correspondence analysis of genes and tissue types and finding genetic links from microarray data. Genome Informatics 11: 83–95. [PubMed] [Google Scholar]
- Kistler HC, Rep M, Ma L‐J. 2013. Structural dynamics of Fusarium genomes In: Brown DW, Proctor RH, eds. Fusarium: genomics, molecular and cellular biology. Norfolk, UK: Caister Academic Press, 31–41. [Google Scholar]
- Kumar L, Breakspear A, Kistler C, Ma LJ, Xie X. 2010. Systematic discovery of regulatory motifs in Fusarium graminearum by comparing four Fusarium genomes. BMC Genomics 11: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Park C, Kim JC, Kim JE, Lee YW. 2010. Identification and functional characterization of genes involved in the sexual reproduction of the ascomycete fungus Gibberella zeae . Biochemical and Biophysical Research Communications 401: 48–52. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar‐Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I et al 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae . Science 298: 799–804. [DOI] [PubMed] [Google Scholar]
- Leslie JF, Summerell BA. 2006. Fusarium laboratory manual. Ames, IA, USA: Blackwell Publishing. [Google Scholar]
- Li YM, Wang CF, Liu WD, Wang GH, Kang ZS, Kistler HC, Xu JR. 2011. The HDF1 histone deacetylase gene is important for conidiation, sexual reproduction, and pathogenesis in Fusarium graminearum . Molecular Plant–Microbe Interactions 24: 487–496. [DOI] [PubMed] [Google Scholar]
- Lysoe E, Seong KY, Kistler HC. 2011. The transcriptome of Fusarium graminearum during the infection of wheat. Molecular Plant–Microbe Interactions 24: 995–1000. [DOI] [PubMed] [Google Scholar]
- Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B et al 2010. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium . Nature 464: 367–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman I, Regev A, Friedman N. 2004. Inferring quantitative models of regulatory networks from expression data. Bioinformatics 20(Suppl 1): i248–i256. [DOI] [PubMed] [Google Scholar]
- Nasmith CG, Walkowiak S, Wang L, Leung WW, Gong Y, Johnston A, Harris LJ, Guttman DS, Subramaniam R. 2011. Tri6 is a global transcription regulator in the phytopathogen Fusarium graminearum . PLoS Pathogens 7: e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N, May GS. 2001. The molecular mechanisms of conidial germination. FEMS Microbiology Letters 199: 153–160. [DOI] [PubMed] [Google Scholar]
- Park B, Park J, Cheong K‐C, Choi J, Jung K, Kim D, Lee Y‐H, Ward TJ, O'Donnell K, Geiser DM et al 2011. Cyber infrastructure for Fusarium: three integrated platforms supporting strain identification, phylogenetics, comparative genomics and knowledge sharing. Nucleic Acids Research 39(Suppl 1): D640–D646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Kim S et al 2008. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics 24: 1024–1025. [DOI] [PubMed] [Google Scholar]
- Pe'er D, Regev A, Elidan G, Friedman N. 2001. Inferring subnetworks from perturbed expression profiles. Bioinformatics 17(Suppl 1): S215–S224. [DOI] [PubMed] [Google Scholar]
- Pe'er D, Regev A, Tanay A. 2002. Minreg: inferring an active regulator set. Bioinformatics 18(Suppl 1): S258–S267. [DOI] [PubMed] [Google Scholar]
- Pe'er D, Tanay A, Regev A. 2006. MinReg: a scalable algorithm for learning parsimonious regulatory networks in yeast and mammals. Journal of Machine Learning Research 7: 167–189. [Google Scholar]
- Qi W, Kwon C, Trail F. 2006. Microarray analysis of transcript accumulation during perithecium development in the filamentous fungus Gibberella zeae (anamorph Fusarium graminearum). Molecular Genetics and Genomics 276: 87–100. [DOI] [PubMed] [Google Scholar]
- Ramachandra S, Linde J, Brock M, Guthke R, Hube B, Brunke S. 2014. Regulatory networks controlling nitrogen sensing and uptake in Candida albicans . PLoS ONE 9: e92734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Kistler HC. 2010. The genomic organization of plant pathogenicity in Fusarium species. Current Opinion in Plant Biology 13: 420–426. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Güldener U, Mannhaupt G, Münsterkötter M et al 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Research 32: 5539–5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz J, Navarro J, Arbués A, Martín C, Marijuán PC, Moreno Y. 2011. The transcriptional regulatory network of Mycobacterium tuberculosis . PLoS ONE 6: e22178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer J, Strimmer K. 2005a. An empirical Bayes approach to inferring large‐scale gene association networks. Bioinformatics 21: 754–764. [DOI] [PubMed] [Google Scholar]
- Schafer J, Strimmer K. 2005b. A shrinkage approach to large‐scale covariance matrix estimation and implications for functional genomics. Statistical Applications in Genetics and Molecular Biology 4: Article 32. [DOI] [PubMed] [Google Scholar]
- Segal E, Shapira M, Regev A, Pe'er D, Botstein D, Koller D, Friedman N. 2003. Module networks: identifying regulatory modules and their condition‐specific regulators from gene expression data. Nature Genetics 34: 166–176. [DOI] [PubMed] [Google Scholar]
- Seong KY, Pasquali M, Zhou X, Song J, Hilburn K, McCormick S, Dong Y, Xu JR, Kistler HC. 2009. Global gene regulation by Fusarium transcription factors Tri6 and Tri10 reveals adaptations for toxin biosynthesis. Molecular Microbiology 72: 354–367. [DOI] [PubMed] [Google Scholar]
- Seong KY, Zhao X, Xu JR, Guldener U, Kistler HC. 2008. Conidial germination in the filamentous fungus Fusarium graminearum . Fungal Genetics and Biology 45: 389–399. [DOI] [PubMed] [Google Scholar]
- Shmulevich I, Dougherty ER, Kim S, Zhang W. 2002. Probabilistic Boolean networks: a rule‐based uncertainty model for gene regulatory networks. Bioinformatics 18: 261–274. [DOI] [PubMed] [Google Scholar]
- Sikhakolli UR, Lopez‐Giraldez F, Li N, Common R, Townsend JP, Trail F. 2012. Transcriptome analyses during fruiting body formation in Fusarium graminearum and Fusarium verticillioides reflect species life history and ecology. Fungal Genetics and Biology 49: 663–673. [DOI] [PubMed] [Google Scholar]
- Son H, Seo YS, Min K, Park AR, Lee J, Jin JM, Lin Y, Cao P, Hong SY, Kim EK et al 2011. A phenome‐based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathogens 7: e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban M, Mott E, Farley T, Hammond‐Kosack K. 2003. The Fusarium graminearum MAP1 gene is essential for pathogenicity and development of perithecia. Molecular Plant Pathology 4: 347–359. [DOI] [PubMed] [Google Scholar]
- Vignes M, Vandel J, Allouche D, Ramadan‐Alban N, Cierco‐Ayrolles C, Schiex T, Mangin B, de Givry S. 2011. Gene regulatory network reconstruction using Bayesian networks, the Dantzig Selector, the Lasso and their meta‐analysis. PLoS ONE 6: e29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang S, Hou R, Zhao Z, Zheng Q, Xu Q, Zheng D, Wang G, Liu H, Gao X et al 2011. Functional analysis of the kinome of the wheat scab fungus Fusarium graminearum . PLoS Pathogens 7: e1002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkopp PJ, Kalay G. 2012. Cis‐regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nature Reviews Genetics 13: 59–69. [DOI] [PubMed] [Google Scholar]
- Wong P, Walter M, Lee W, Mannhaupt G, Munsterkotter M, Mewes HW, Adam G, Guldener U. 2011. FGDB: revisiting the genome annotation of the plant pathogen Fusarium graminearum . Nucleic Acids Research 39: D637–D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu HY, Seo JA, Kim JE, Han KH, Shim WB, Yun SH, Lee YW. 2008. Functional analyses of heterotrimeric G protein G alpha and G beta subunits in Gibberella zeae . Microbiology 154: 392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Townsend JP. 2010. The filamentous fungal gene expression database (FFGED). Fungal Genetics and Biology 47: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Waalwijk C, de Wit P, Tang D, van der Lee T. 2014. Relocation of genes generates non‐conserved chromosomal segments in Fusarium graminearum that show distinct and co‐regulated gene expression patterns. BMC Genomics 15: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XM, Zhang XW, Tang WH, Chen L. 2009. FPPI: Fusarium graminearum protein–protein interaction database. Journal of Proteome Research 8: 4714–4721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1 Scatterplot matrix of three biological replicates and associated Pearson correlation coefficients (r) for each condition/biological state transcriptomically profiled in this study using the Affymetrix Fungal Multigenome ExonChip.
Fig. S2 Bayesian network score distribution for the top 300 regulators in the Fusarium graminearum gene regulatory network (GRN).
Fig. S3 Visualization of the Fusarium graminearum gene regulatory network (GRN) featuring the top 120 regulators, including transcription factors (TFs) and signal proteins (SPs), represented by red and green nodes, respectively.
Fig. S4 Gene–phenotype networks depicting the association of known phenotypes (red nodes) with inferred signal protein (SP) (blue nodes) and transcription factor (TF) (green nodes) regulators.
Fig. S5 Regulatory modules are differentially regulated in response to various biological conditions.
Fig. S6 Heatmap of hierarchical co‐clustering of biological conditions (rows) and 120 regulators (columns).
Fig. S7 Biased regulatory networks for core regulators of Fusarium graminearum.
Fig. S8 Shared networks by the Fusarium graminearum gene regulatory network (GRN) and protein–protein interaction network (FPPI).
Table S1 Summary of conditions on which the transcriptomic data are based for Fusarium graminearum gene regulatory network inference in this study
Table S4 Summary of signal proteins identified as inferred regulators in this network
Table S2 Input gene expression data matrix
Table S5 List of 75 inferred transcription factor regulators and the most enriched binding sites for target genes
Table S8 Shared networks by the Fusarium graminearum gene regulatory network (GRN) and protein–protein interaction network (FPPI)
Table S3 Spreadsheets containing lists of candidate regulators, inferred 120 top regulators, the full gene regulatory network (GRN), core genome GRN and Fusarium graminearum‐specific GRN
Table S6 Gene lists of eight regulatory modules (A–H) and complete FunCat analysis results
Table S7 Gene regulatory networks for candidate effector genes


