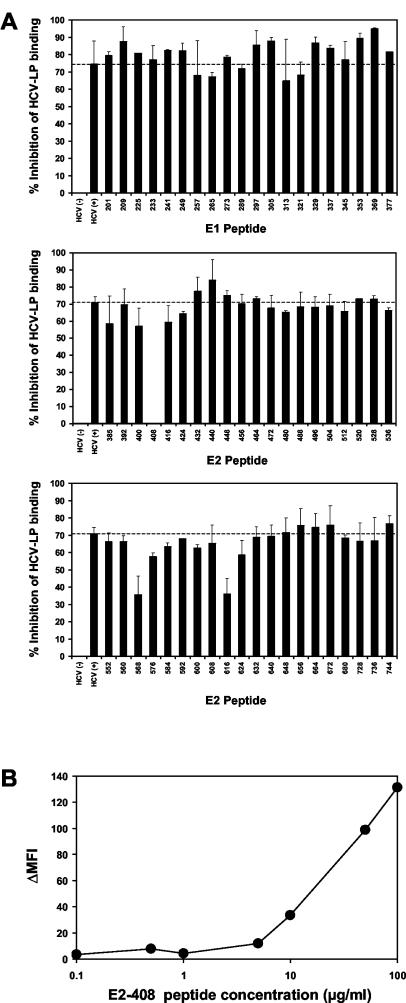
FIG. 6.
Epitope mapping of antibodies inhibiting cellular binding of HCV-LPs. (A) To identify HCV-LP envelope epitopes targeted by human antibodies inhibiting cellular HCV-LP binding, an anti-HCV-positive serum (HCV+) with marked inhibition of binding activity (Fig. 3) was incubated with overlapping 15-mer peptides of the HCV envelope glycoproteins (comprising amino acid positions 201 to 758) derived from the HCV-J strain (26) or PBS (as a control) for 1 h at RT (peptide concentration, 100 μg/ml; serum dilution, 1:50). HCV-LPs (derived from the homologous isolate as the peptides) were then added to the peptide-antibody complexes and incubated for 1 h at 37°C. The HCV-LPs, antibodies, and peptides were added to HuH-7 cells and incubated for 1 h at 4°C. Following the removal of nonbound HCV-LPs, antibodies, and peptides by washing the cells in PBS, binding of HCV-LPs was detected by flow cytometry as described above. Inhibition of HCV-LP binding (as indicated on the y axis) was calculated as described in the legend to Fig. 2. The error bars indicate standard deviations of results of a representative experiment performed in triplicate. (B) Concentration-dependent reversion of HCV-LP inhibition of binding activity by E2 peptide 408. Anti-HCV-positive serum (dilution, 1:50) was preincubated with E2 peptide 408 (SQKIQLVNTNGSWHI; corresponding to amino acid positions 408 to 422) at the peptide concentrations indicated on the x axis. Cellular HCV-LP binding (corresponding to ΔMFI, indicated on the y axis) in the presence of the antiviral antibody-peptide complex was determined as described in Materials and Methods.