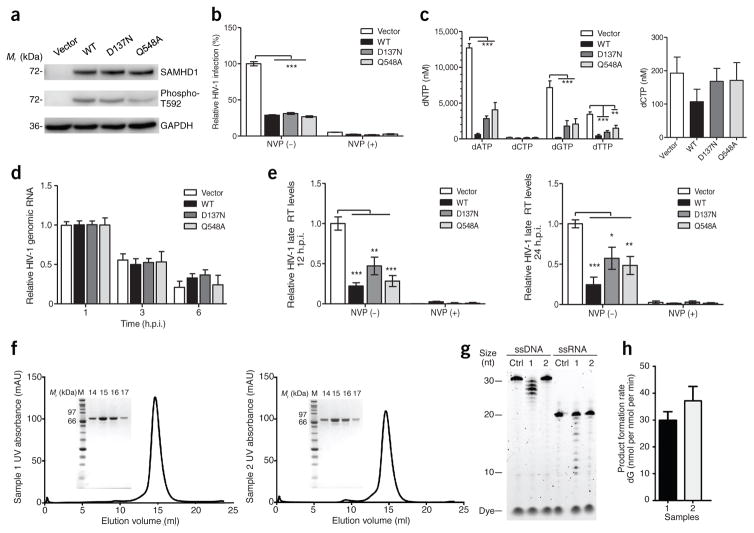
Figure 1.
SAMHD1-mediated HIV-1 restriction in cells does not involve RNase activity. (a–e) Both D137N and Q548A mutants of SAMHD1 restrict HIV-1 infection in PMA-differentiated U937 cells by decreasing viral cDNA synthesis, but not viral genomic RNA (gRNA). U937 cell lines stably expressing WT and mutant SAMHD1 were differentiated with PMA (30 ng/ml) for 20 h. The reverse transcriptase inhibitor nevirapine (NVP) was used as a control. (a) Lysates from PMA-differentiated cells were collected for immunoblotting to confirm SAMHD1 expression and T592-phosphorylated SAMHD1 (phospho-T592). GAPDH was used as a loading control. Blot is representative of three independent experiments. (b) PMA-differentiated cells were infected with single-cycle HIV-1-Luc/VSV-G at a multiplicity of infection (MOI) of 1. At 24 h postinfection (h.p.i.), luciferase assay was used to measure HIV-1 infectivity. Graph depicts relative percentage of luciferase per 10 μg of protein, with the vector set as 100%. Data are presented as mean ± s.e.m. of six independent experiments with two biological replicates per experiment. Statistical analysis was performed using the one-way ANOVA with Dunnett’s correction, ***P < 0.001. (c) Decreased dNTP levels of PMA-differentiated U937 cell lines expressing WT or mutant SAMHD1. Data are presented as means ± s.e.m. of two independent experiments with two biological replicates per experiment. The average concentrations of dNTPs (left) and dCTP (right) in different cell lines are shown. Statistical analysis was performed using the Kruskal–Wallis one-way ANOVA. (d) SAMHD1 does not degrade HIV-1 gRNA in PMA-differentiated U937 cells. The levels of HIV-1 gRNA were measured at 1, 3 and 6 h.p.i., as described at an MOI of 1. Data are presented as mean ± s.e.m. of three independent experiments with two biological replicates per experiment and depicted as relative to HIV-1 gRNA, with the time point 1 h.p.i. set as 1. RNA samples without reverse transcription were used as negative controls and showed no detection of HIV-1 gRNA (data not shown). (e) The expression of WT or mutant SAMHD1 reduces HIV-1 late reverse transcription (RT) products in PMA-differentiated U937 cells at 12 and 24 h.p.i. The levels of HIV-1 late RT products were measured by qPCR. Data are presented as means ± s.e.m. (n = 4 experiments for 12 h.p.i. and n = 6 experiments for 24 h.p.i.). Statistical analysis was performed using the one-way ANOVA with Dunnett’s correction, ***P < 0.001, **P < 0.01, *P < 0.05. (f–h) Characterization of SAMHD1 enzymatic activities in vitro. (f) The purity of recombinant WT SAMHD1 from different batches of preparations (sample 1 and sample 2), as demonstrated by analytical size-exclusion chromatography and SDS–PAGE. SAMHD1 (150 μl) at 2 mg/ml was applied to a Superdex 200 10/300 GL column. Fraction numbers on the SDS–PAGE indicate elution volumes. (g) Moderate exonuclease activities on both ssDNA and ssRNA were observed for sample 1; only background-level activities were detected for sample 2. Ctrl indicates a control without protein. The nuclease-activity assays were performed at 37 °C for 1 h, with 1 μM of SAMHD1 and 1 μM of ssDNA or ssRNA in the presence of 5 mM Mg2+. (h) The dNTPase activity of SAMHD1 proteins (0.5 μM) was assayed with 1 mM dGTP from 5–15 min. The amount of dG products generated in the reactions was quantified by HPLC. Error bars represent s.e.m. from triplicate experiments.