FIG 2 .
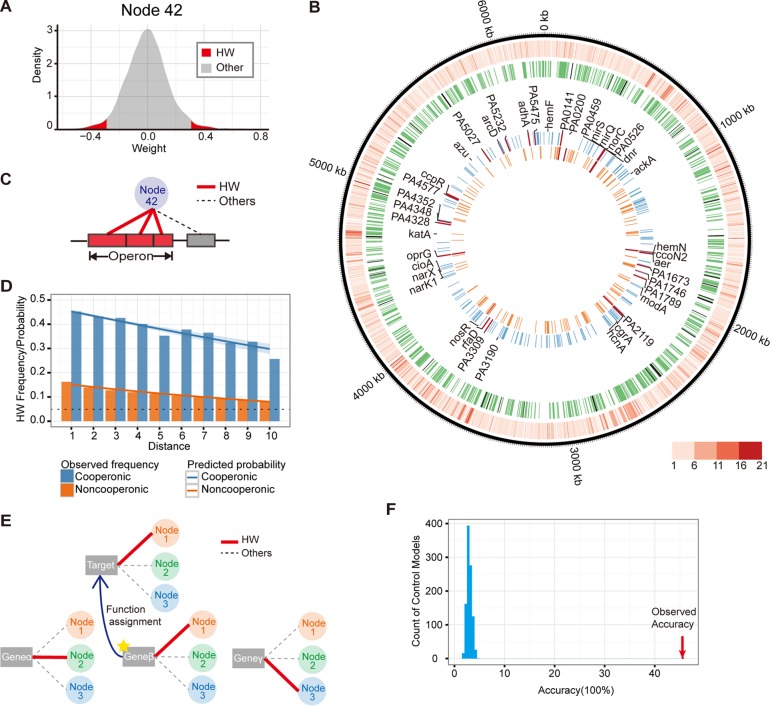
ADAGE weights reflect a gene’s common regulatory and process features. (A) HW genes defined in ADAGE. The distribution of edge weights that connect genes to each node, e.g., node 42 shown here, is approximately normal. HW genes in a node were defined as genes whose weights were more than 2 standard deviations from the mean (shown in red). (B) The contributions of individual genes and operons to the ADAGE model. The outermost ring shows those genes that are HW within at least one node. The color intensity reflects how many nodes (ranging from 1 to 21) a gene was connected to at a high weight. The second-to-outermost circle shows operons that were significantly associated with at least one node (green bars) and those that were not significantly associated with any of the 50 nodes (black bars). The inner two circles represent the HW genes in nodes 42 and 30 (outside to inside). The labeled genes are those identified by Trunk et al. (37) and Jackson et al. (36) as regulated by Anr and they are colored red among HW genes of nodes 30 or 42. The HW genes in node 42 overlapped extensively with Anr-regulated genes, which suggested that node 42 captured the regulatory signature of Anr. (C and D) ADAGE captured principles of bacterial genome organization. (C) In the bacterial genome, genes are arranged into operons, which the ADAGE model recognizes by connecting cooperonic genes to a shared node. (D) The regulatory role of genome positioning in P. aeruginosa was captured by ADAGE. A logistic regression analysis revealed that cooperonic genes (blue line shows model data; bars show observed data) were more likely to be co-HW genes than noncooperonic genes (red). As the number of genes between two genes on the chromosome increased, they were less likely to be co-HW. The black dotted line represents the background frequency of HW genes across all nodes. (E and F) ADAGE captured gene functional features. (E) We found the closest neighbor of a target gene based on the Euclidean distance between the weight vectors connecting each gene to 50 nodes and assigned the closest neighbor gene’s function to the target gene. (F) The accuracy of gene function assignment using the ADAGE model (45%, red arrow) was much higher than the accuracies achieved with 1,000 randomly permuted control models (distribution shown in blue). Here, we considered a function assignment positive if one or more functions assigned by the closest neighbor gene matched the target gene’s annotations.