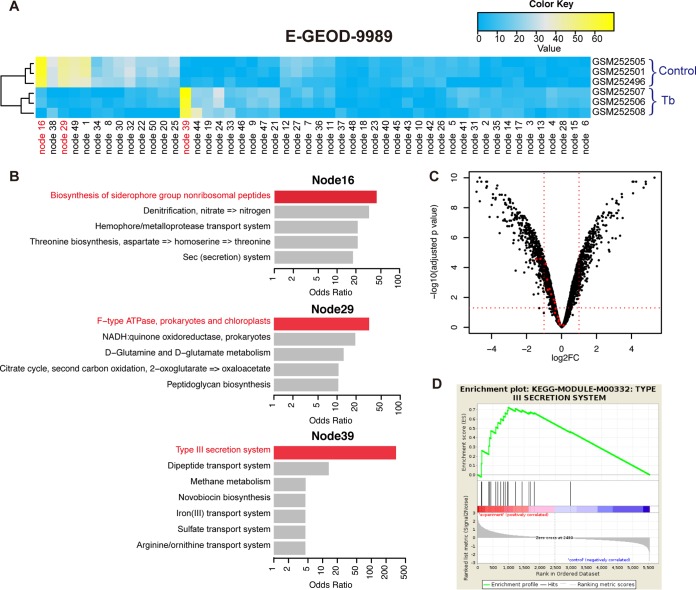
FIG 5 .
Reanalysis of an existing experiment using ADAGE. (A) Node activity heat map for the data set E-GEOD-9989. Nodes 39, 16, and 29 are the three nodes that most strongly differentiated control samples from those challenged with tobramycin. (B) KEGG pathway analysis of differentially active nodes. The top 5 most enriched KEGG pathways (including ties) based on odds ratios are shown for each node. Nodes 39, 16, and 29 each represent a cellular process being influenced in the experiment (in red), especially node 39, which captures the subtle change in the T3SS pathway. (C) Volcano plot showing differentially expressed genes in E-GEOD-9989. The horizontal dotted red line indicates an adjusted P value cutoff of 0.05, and the two vertical lines correspond to log2 fold changes of −1 and 1. Genes in the KEGG T3SS pathway (KEGG-Module-M00332) are highlighted in red. Although only two genes passed both the significance cutoff and fold change cutoff, genes in the pathway showed consistently lower expression in tobramycin-treated samples. (D) Gene set enrichment analysis of the T3SS pathway. GSEA also captured the consistent differential expression pattern of T3SS, which it ranked as the 12th most significantly enriched pathway.