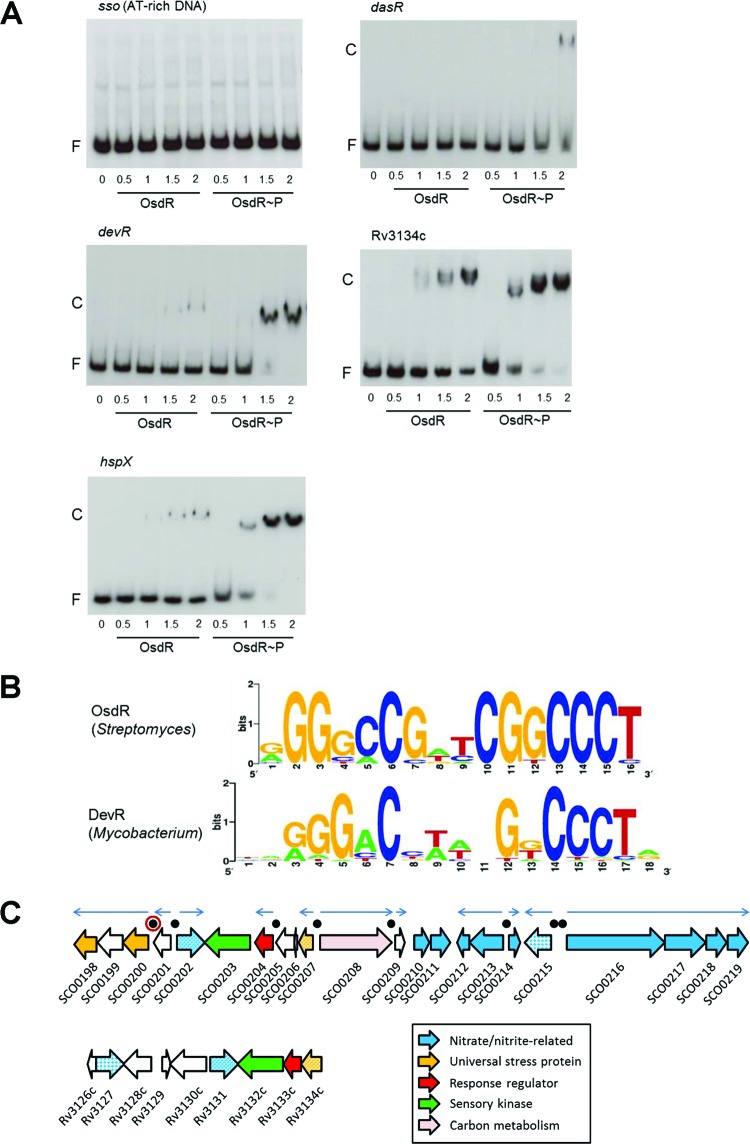
FIG 2 .
Binding site of OsdR (SCO0204) and comparison to that of M. tuberculosis DevR. (A) EMSAs using purified His6-tagged OsdR on known targets of DevR in M. tuberculosis. Both phosphorylated OsdR (OsdR~P) and nonphosphorylated OsdR (OsdR) were used in the assays. OsdR~P was obtained after AcP phosphorylation in vitro. Numbers on the horizontal axis refer to micromolar concentrations. F, free DNA fragment; C, complexes of DNA and protein. (B) Weblogo representation of cis-regulatory elements identified upstream of osdR, identified from the upstream regions of osdR orthologues in 12 Streptomyces species (see Materials and Methods). For comparison, the upstream regulatory element recognized by DevR (97) is presented. (C) Gene synteny between the loci around osdR in S. coelicolor (top) and devR in M. tuberculosis (bottom). Functional categories are given in the figure, and black dots indicate predicted OsdR binding sites, with that of upsA (SCO0200) surrounded by a red ring. Orthologues are presented in the same colors, and when multiple genes with similar functions are present, they appear in patterns.