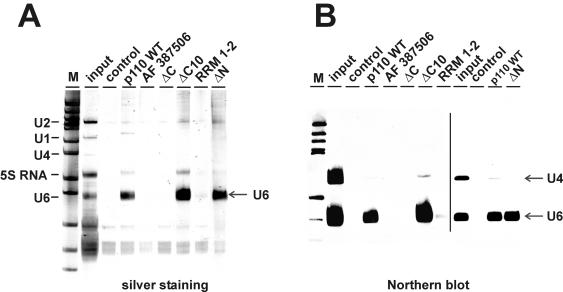
FIG. 2.
Mutational analysis of p110 protein-U6 snRNA binding in vitro. Total RNA from a HeLa cell S100 extract was incubated with recombinant full-length p110 protein or an equimolar amount of splice variant AF 387506 or mutant derivative ΔC, ΔC10, RRM 1-2, or ΔN. Ten percent of the RNA input was analyzed in parallel (input lanes). Control reactions were carried out in the absence of added p110 protein (control lanes). The p110-RNA complexes formed were immunoprecipitated with anti-p110 antibodies, and the coprecipitated RNAs were analyzed by denaturing gel electrophoresis and detected by silver staining (A) or by Northern blotting with U4- and U6-specific probes (B). The line in panel B separates autoradiograms from two separate experiments. The mobilities of the U1, U2, U4, U6, and 5S RNAs are marked on the left. Lanes M, pBR322/HpaII marker fragments (A) and DIG marker V (Roche) (B). The arrows indicate the positions of the U4 and U6 snRNAs.