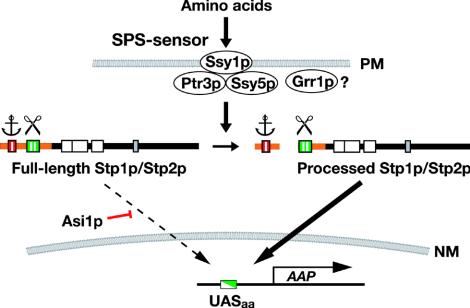
FIG. 7.
Model of the amino acid-induced SPS-sensing pathway. The latent nature of Stp1p/Stp2p is determined by conserved sequence motifs region I (red) and region II (green) present in their N-terminal domains (orange). Region I appears to function as a cytoplasmic retention signal (anchor) that prevents the unprocessed full-length forms from efficiently entering the nucleus. Region II is required for amino acid-induced endoproteolytic processing (scissor). In the absence of Asi1p, full-length Stp1p/Stp2p can enter the nucleus at rates sufficient to partially derepress amino acid permease (AAP) gene expression. Extracellular amino acids induce the proteolytic processing of Stp1p/Stp2p in an SPS sensor- and Grr1p-dependent manner. The precise function of Grr1p remains to be defined (?); Grr1p either functions concomitantly with the previously characterized SPS sensor components (Ssy1p-Ptr3p-Ssy5p) or is required for SPS sensor assembly. The processed shorter forms of Stp1p and Stp2p containing DNA-binding domains (white boxes) accumulate in the nucleus, where they bind UAS present within the promoters of amino acid permease genes (UASaa). PM, plasma membrane; NM, nuclear membrane.