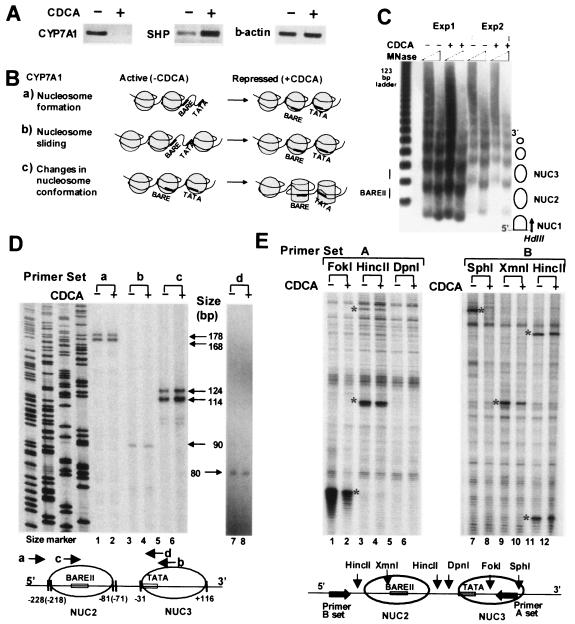
FIG. 1.
Bile acid-induced chromatin remodeling at the CYP7A1 promoter. (A) Effects of bile acid treatment on SHP and CYP7A1 expression. Total RNA was isolated from HepG2 cells and subjected to semiquantitative RT-PCR using primer sets specific for human CYP7A1, SHP, and β-actin. +, present; −, absent. (B) Three potential models of chromatin remodeling after CDCA treatment. (C) Low-resolution mapping of nucleosome positions. Permeabilized HepG2 cells (Exp1) or isolated nuclei (Exp2) were incubated with MNase, and purified DNA was cleaved with HindIII and subjected to indirect end-labeling genomic Southern analysis using the probe indicated by the arrow. Nucleosome positions are indicated as circles. (D) High-resolution mapping of nucleosome positions. HepG2 cells were incubated with formaldehyde to cross-link proteins and nucleic acids. Nuclei were isolated and partially digested with MNase. After the cross-links were reversed, nucleosomal DNA was isolated and subjected to LMPCR using the primer sets a, b, c, and d. Sequencing ladders served as size markers. The lengths of LMPCR products are indicated. A schematic diagram of nucleosome positioning at the promoter is shown below. (E) Accessibility of CYP7A1 chromatin to restriction endonucleases. Isolated nuclei were partially digested with restriction enzymes. DNA was purified and subjected to LMPCR using primer set A or B. The asterisks indicate the restriction enzyme cleavages detected by LMPCR. Background bands present in all lanes result from nonspecific cleavages by endogenous nucleases. Reproducible results were obtained in two to five independent studies.