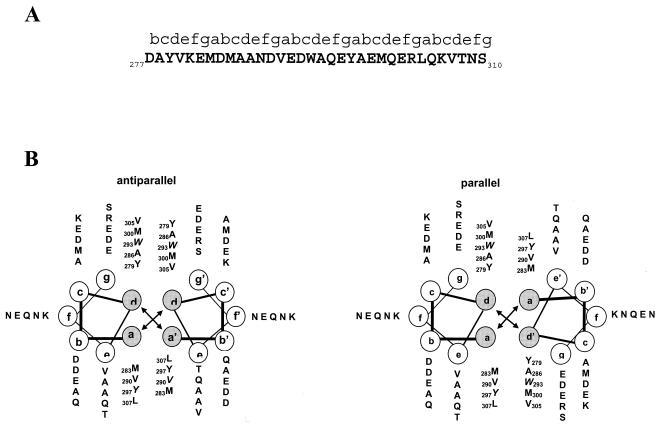
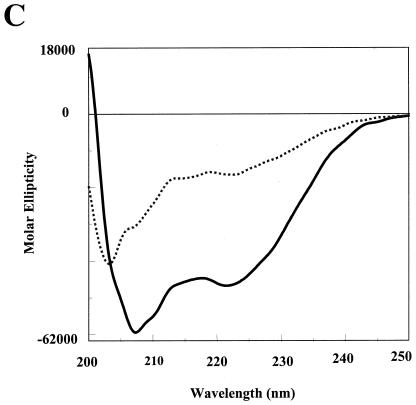
FIG. 2.
In silico analysis of the LdPEX5 coiled coil. (A) Primary sequence for the predicted LdPEX5 coiled coil (residues 277 to 310). Above the amino acid sequence is the designation of the heptad repeats using the abcdefg nomenclature. (B) The hydrophobic core contacts, a↔a′ and d↔d′ for the parallel orientation or the a↔d′ and a′↔d for the antiparallel arrangement, that stabilize the coiled-coil homodimer packing are illustrated by plotting the primary sequence on helical wheel diagrams. (C) The secondary structure of ldpex5 268-303 was determined by circular dichroism on a JASCO 710 spectropolarimeter with a 0.1-cm cylindrical quartz cuvette. Peptide was dissolved at a concentration of 1 mg/ml in 40 mM sodium phosphate 100 mM NaCl (pH 7.2; dashed line) or 50% trifluoroethanol-40 mM sodium phosphate-100 mM NaCl (pH 7.2; solid line). Spectra represent the average of 10 scans from 200 to 250 nm, and the buffer baseline has been subtracted from each spectra. The uncorrected percent helicity was calculated with the [θ]222 by the method of Wu et al. (55) using the following equation: % helix = {([θ]222 + 2,000)/(−37,400 + 2,000)} × 100.