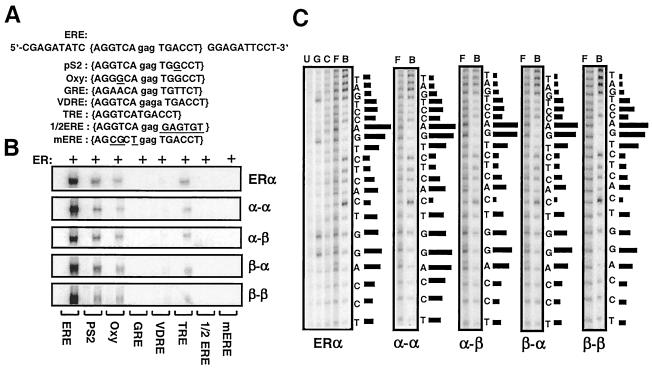
FIG. 5.
(A) Upper strand of the sequence containing the consensus ERE (in brackets), pS2, and Oxy ERE, GRE, VDRE, and TRE responsive elements and 1/2 ERE and mERE with the underlined five- and three-nucleotide changes from the ERE, respectively. The central base spacer is shown in lowercase. (B) Fusion ERs bind to the same spectrum of response elements. Equal molar concentrations of in vitro-synthesized ERα dimer or a fusion receptor were subjected to EMSA. A representative phosphorimage of two independent experiments is shown. (C) Fusion receptors bind to ERE in a manner similar to ERα. Critical nucleosides for ER-ERE interaction were identified by missing nucleoside hydroxyl radical assay. End-labeled DNA fragment containing consensus ERE was randomly cleaved by hydroxyl radical, incubated with 50 μl of transcription-translation mixtures, and subjected to 5% nondenaturing PAGE. Radioactive bands containing bound and free DNA were excised, eluted, and subjected to 15% sequencing gel electrophoresis. The intensities of individual DNA bands from sequencing gels were quantified by using a PhosphorImager. The ratio of free (F) to bound (B) DNA at each base was plotted as horizontal bars, the length of which approximates the strength of nucleoside contact with the protein. Uncut (U), Maxam-Gilbert G reaction-cut (G), and randomly cut (C) DNA fragments were also electrophoresed. A representative phosphorimage from at least two independent experiments is shown.