Fig. 2.
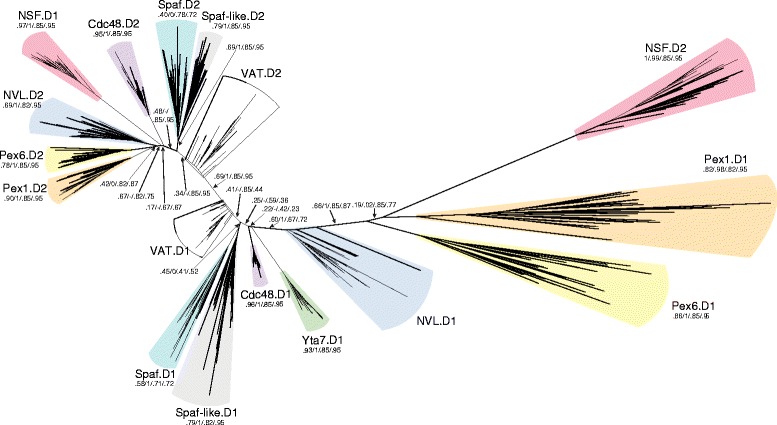
Evolutionary tree of the individual AAA domains of the different Cdc48 family members. The tree was constructed from the individual AAA domains (D1- and D2-domains) of the Cdc48 family using a selection of 48 eukaryotic and 26 archaeal species, accounting for a total of 687 AAA domains. The representative species are listed in Additional file 2: Table S2. The different family members are highlighted in different colors, while D1- and D2-domains of the same protein have the same color. Statistical support values (likelihood-mapping/IQ-TREE support/RAxML support/PhyML support) are given at selected inner edges. Most AAA domains form short branches and split into a D1 and a D2 subtree, in which the two domains of all archaeal Cdc48 are located close to the center of the tree, probably reflecting the fact that the eukaryotic family members are derived from a primordial VAT [19]. However, in the tree all archaea sequences, even from the recently found Lokiarchaeota, are well-separated from eukaryotic family members. The two more divergent D1-domains of the peroxins and the D2 domain of the N-ethylmaleimide sensitive factor (NSF) form long branches. Notably, the D2-domain of NSF is located in the D1 subtree, whereas the D1-domain of NSF is found in the D2 subtree. A similar pattern had been observed in earlier studies and it has been suggested that the two domains of NSF have been swapped during evolution [118]. Given the generally conserved architecture of the protein family, this scenario is not very likely [39]. It is much more probable that this branching pattern is caused by long-branch attraction. In fact, when we included the incomplete, long-branching D2-domain of Yta7, the branching pattern of the two NSF domains changed (Additional file 11: Figure S7)
