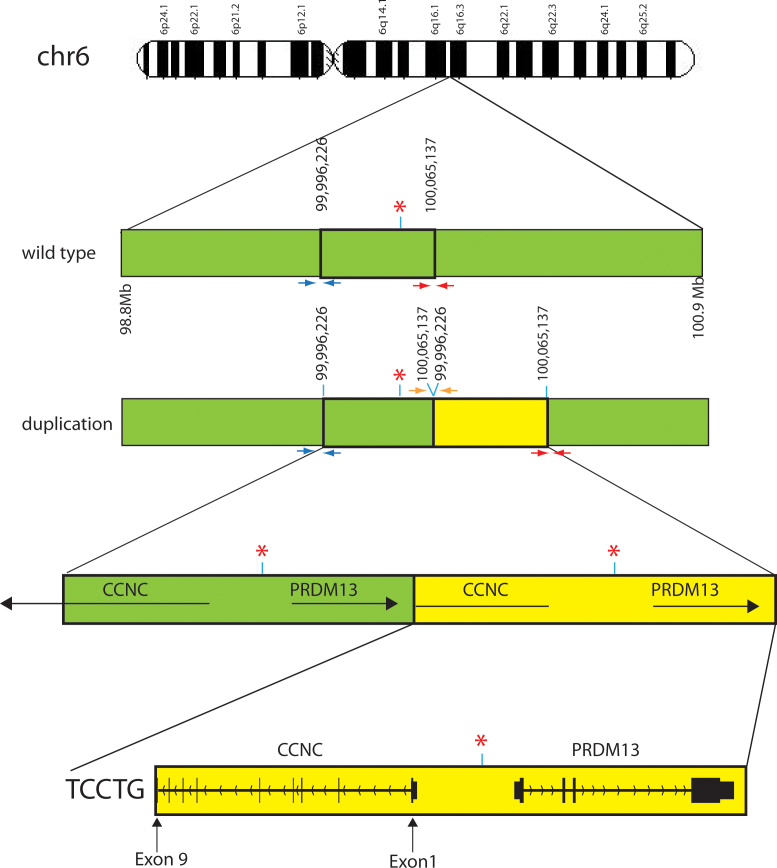
Figure 3.
Pictorial representation of the wild-type and pathogenic duplicated regions on chromosome 6. Green bars indicate the wild-type region on chromosome 6 while the yellow bars indicate the duplicated region. Red asterisks indicate the DNase I hypersensitivity site. Small arrows below the chromosome bar pictorials are the three pairs of PCR primers that produce different size products. The PCR product generated from these primers was used to determine the presence or absence of the deletion and the exact sequence of the duplication in family members (Figure 1B). Red and blue primers should amplify from the wild-type and duplicated chromosomes. The orange primers amplify only on the duplicated chromosome. Sequencing of the PCR product resulting from the orange depicted primers determined the exact duplication break point and the inclusion of 5 bp of exogenous DNA, TCCTG, between the wild-type and duplicated regions. Large horizontal black arrows with gene names above illustrate the strand and transcriptional direction of each gene. Vertical lines and blocks represent exons. The duplication found of the CCNC gene is only from exons 1 through 9 making it unlikely that any protein is generated from the duplicated region. The presence of the entire PRDM13 gene downstream from the DNase hypersensitivity site likely causes a change in PRDM13 protein levels compared to wild-type levels.