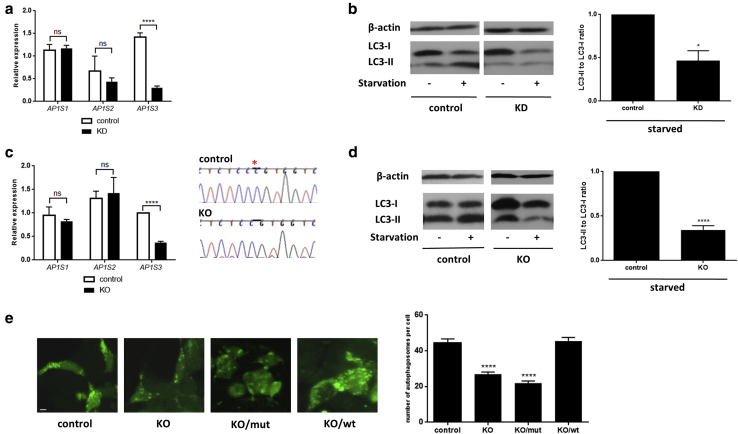
Figure 2.
AP1S3 deficiency results in impaired autophagy. (a) After the generation of a HaCaT AP1S3 knockdown cell line, gene silencing was measured by real-time PCR, because of cross-reactivity of the anti-AP1σ1c antibody with the proteins encoded by AP1S1 and AP1S2. (b) Starvation-induced LC3-II accumulation was measured by Western blotting and densitometry. The data are presented as mean ± standard error of the mean of measurements obtained in four independent experiments. (c) HEK293 AP1S3 knockout cell lines harboring a c.124delC change (highlighted by a red asterisk in the chromatogram) were generated by CRISPR/Cas-9 editing. (d) Cells were starved to induce autophagy, and LC3-II accumulation was measured by Western blotting. The data are presented as described. (e) Control and AP1S3 KO HEK293 cells were transfected with GFP-LC3 and either an empty vector (control and KO panels) or a rescue construct (wild-type AP1S3 in KO/wt panel and p.Arg33Trp AP1S3 in KO/mut). Starvation-induced LC3 punctae were visualized by confocal fluorescence microscopy. The data are presented as mean ± standard error of the mean of measurements obtained in at least 15 cells per experiment. Scale bar = 5 μm. ∗P ≤ 0.05, ∗∗∗∗P ≤ 0.0001. Cas9, CRISPR-associated protein-9; CRISPR, clustered regularly interspaced short palindromic repeats; GFP, green fluorescent protein; KD, knockdown; KO, knockout; mut, mutated; ns, not significant; wt, wild-type.